 | ||
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity (e.g., self-splicing) can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing (see spliceosome) may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically mobilize to new DNA sites has been exploited as a tool for biotechnology.
Contents
Structure and catalytic site
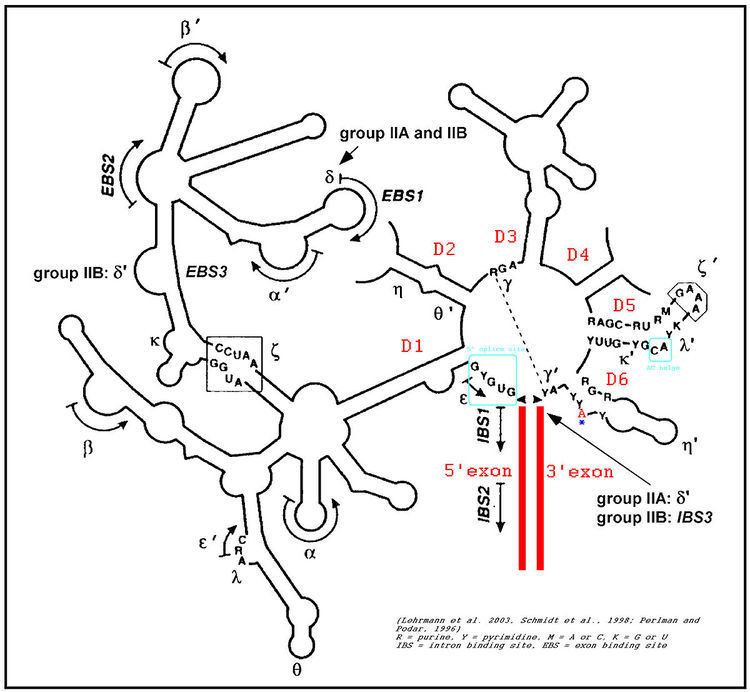
The secondary structure of group II introns is characterized by six typical stem-loop structures, also called domains I to VI or D1 to D6. The domains radiate from a central core that brings the 5' and 3' splice junctions into close proximity. The proximal helix structures of the six domains are connected by a few nucleotides in the central region (linker or joiner sequences). Due to its enormous size, the domain 1 was divided further into subdomains a, b, c, and d. Sequence differences of group II introns that led to a further division into subgroups IIA and IIB were identified. Group II introns also form very complicated RNA Tertiary Structure.
Group II introns possess only a very few conserved nucleotides, and the nucleotides important for the catalytic function are spread over the complete intron structure. The few strictly conserved primary sequences are the consensus at the 5' and 3' splicing site (...↓GUGYG&... and ...AY↓...), some of the nucleotides of the central core (joiner sequences), a relatively high number of nucleotides of D5 and some short-sequence stretches of D1. The unpaired adenosine in D6 marked by an asterisk (7 or 8 nt away from the 3' splicing site, respectively) is also conserved and plays a central role in the splicing process.
In 2005, A. De Lencastre et al. found that during splicing of Group II introns, all reactants are preorganized before the initiation of splicing. The branch site, both exons, the catalytically essential regions of D5 and J2/3, and epsilon−epsilon' are in close proximity before the first step of splicing occurs. In addition to the bulge and AGC triad regions of D5, the J2/3 linker region, the epsilon−epsilon' nucleotides and the coordination loop in D1 are crucial for the architecture and function of the active-site.
Group II catalytic intron
Group II catalytic introns are found in rRNA, tRNA, and mRNA of organelles (chloroplasts and mitochondria) in fungi, plants, and protists, and also in mRNA in bacteria. They are large self-splicing ribozymes and have 6 structural domains (usually designated dI to dVI). This model and alignment represents only domains V and VI. A subset of group II introns also encode essential splicing proteins in intronic ORFs. The length of these introns can, therefore, be up to 3 kb. Splicing occurs in almost identical fashion to nuclear pre-mRNA splicing with two transesterification steps. The 2' hydroxyl of a bulged adenosine in domain VI attacks the 5' splice site, followed by nucleophilic attack on the 3' splice site by the 3' OH of the upstream exon. Protein machinery is required for splicing in vivo, and long-range intron-intron and intron-exon interactions are important for splice site positioning. Group II introns are further sub-classified into groups IIA and IIB, which differ in splice site consensus, and the distance of the bulged adenosine in domain VI (the prospective branch point forming the lariat) from the 3' splice site.
