 | ||
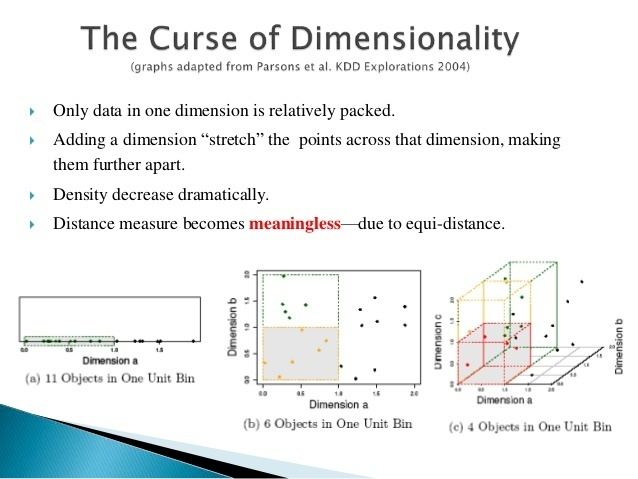
Clustering high-dimensional data is the cluster analysis of data with anywhere from a few dozen to many thousands of dimensions. Such high-dimensional spaces of data are often encountered in areas such as medicine, where DNA microarray technology can produce a large number of measurements at once, and the clustering of text documents, where, if a word-frequency vector is used, the number of dimensions equals the size of the vocabulary.
Contents
Problems
Four problems need to be overcome for clustering in high-dimensional data:
Recent research indicates that the discrimination problems only occur when there is a high number of irrelevant dimensions, and that shared-nearest-neighbor approaches can improve results.
Approaches
Approaches towards clustering in axis-parallel or arbitrarily oriented affine subspaces differ in how they interpret the overall goal, which is finding clusters in data with high dimensionality. An overall different approach is to find clusters based on pattern in the data matrix, often referred to as biclustering, which is a technique frequently utilized in bioinformatics.
Subspace clustering
The image on the right shows a mere two-dimensional space where a number of clusters can be identified. In the one-dimensional subspaces, the clusters
The problem of subspace clustering is given by the fact that there are
Projected clustering
Projected clustering seeks to assign each point to a unique cluster, but clusters may exist in different subspaces. The general approach is to use a special distance function together with a regular clustering algorithm.
For example, the PreDeCon algorithm checks which attributes seem to support a clustering for each point, and adjusts the distance function such that dimensions with low variance are amplified in the distance function. In the figure above, the cluster
PROCLUS uses a similar approach with a k-medoid clustering. Initial medoids are guessed, and for each medoid the subspace spanned by attributes with low variance is determined. Points are assigned to the medoid closest, considering only the subspace of that medoid in determining the distance. The algorithm then proceeds as the regular PAM algorithm.
If the distance function weights attributes differently, but never with 0 (and hence never drops irrelevant attributes), the algorithm is called a "soft"-projected clustering algorithm.
Hybrid approaches
Not all algorithms try to either find a unique cluster assignment for each point or all clusters in all subspaces; many settle for a result in between, where a number of possibly overlapping, but not necessarily exhaustive set of clusters are found. An example is FIRES, which is from its basic approach a subspace clustering algorithm, but uses a heuristic too aggressive to credibly produce all subspace clusters.
Correlation clustering
Another type of subspaces is considered in Correlation clustering (Data Mining).
