 | ||
C6orf222 is a protein that in humans is encoded by the C6orf222 gene (6p21.31). C6orf222 is conserved in mammals, birds and reptiles with the most distant ortholog being the green sea turtle,Chelonia mydas. The C6orf222 protein contains one mammalian conserved domain: DUF3293. The protein is also predicted to contain a BH3 domain, which has predicted conservation in distant orthologs from the clade Aves.
Contents
Gene
C6orf222 is located on the negative DNA strand of the short arm of chromosome 6 at locus 21.31. The gene is 21,129 base pairs long and extends from base pair 36315757 to 36336977. The gene produces a single transcript that is 3,750 base pairs long. There is a unique upstream, in-frame stop codon in the 5’UTR region from base pairs 86-88 of the mRNA.
Gene Neighborhood
The gene PNPLA1 is found on the positive strand just upstream of C6orf222 and belongs to the patatin-like phospholipase family. PNPLA1 extends from base pairs 36239766 to 36313955. ETV7 is found downstream on the negative strand and extends from base pairs 36354221 to 36387800. There are two genes located upstream (LOC105375036) and downstream (LOC105375037) of C6orf222 on the negative and positive strand, respectively. These genes code for Non-coding RNA.
Gene Expression
There are several lines of evidence that suggest the C6orf222 protein is expressed non-ubiquitously in select tissue locations at low levels, including the ascites, bladder, intestine, testes, vagina, lymph nodes and skin. The ascites and bladder displayed the strongest expression with 25 and 66 transcripts per million, respectively.
Promoter
The promoter region for C6orf222 is predicted to exist between 36304561 and 36305162 and has a length of 602 base pairs.
Protein
C6orf222 consists of 652 amino acids, has a weight of 71.9 kDa and an isoelectric point of 8.70 in humans.
Function
While the exact function of C6orf222 remains unknown, there has been evidence for it being involved in apoptosis as a pro-apoptotic protein due to the conserved BH3 interacting domain death agonist located at the C-terminus of the protein between amino acid residues 535 to 611 in humans.
Structure
C6orf222 contains a single domain of unknown function, DUF3292, found between amino acid residues 38-110. The protein is proline rich but asparagine and tyrosine poor. The charge distribution of the protein is fairly equal in that there are no positive or negative charge clusters sequestered within the protein.
The secondary structures of the protein were predicted via multiple bioinformatics servers. All four programs predicted extensive disorder in the protein at the N-terminus, ranging from 79% to 88% of the protein. This extent of disorder is consistent with the amino acid composition of the protein and being proline rich. There were several α-helix structures predicted at the C-terminus of the protein and conserved in orthologs of the human protein. The protein was predominantly soluble, with a majority of the amino acid residues being exposed.15 The average of hydrophobicity for c6orf222 was -0.968, indicating the soluble nature of the protein.
Post-Translational Modifications
There is extensive, predicted phosphorylation of C6orf222, with 42 phosphoserines and 7 phosphothreonines being conserved in orthologs of the human C6orf222 protein. These results implicate C6orf222 as being a phosphoprotein. The protein contains only one nuclear export signal residue, found at 275-L; however, the NES score was rather low at 0.672. Structural analysis of the protein to be sequestered in the nucleus with a 95% probability. This prediction is supported by the presence of a nuclear localization sequence (NLS) found between residues 142-151.
Protein Interactions
Although the bioinformatics programs MINT, STRING and Gene Cards did not reveal any protein interactions with C6orf222. A predicted BH3 domain in the C6orf222 protein was found to interact with both Bcl-2 and Bcl-xL, implicating C6orf222 as being involved in apoptosis.
Table of Potential Transcription Factor Binding Sites in the Predicted C6orf222 Promoter:
Orthologous Space
82 organisms have predicted orthologs with C6orf222. The most distant ortholog is the green sea turtle, which diverged from humans 296 million years ago, indicating C6orf222 developed in reptiles and birds.
Table of C6orf222 Orthologs:
Paralogous Space
There are no predicted paralogs for C6orf222 in both humans and mice.
Conserved Regions
Multiple sequence alignments demonstrated amino acid reside conservation throughout the C6orf222 protein in a variety of orthologs, with the most extensive conservation being found at the C-terminus of the protein.The BH3 interacting-domain death agonist (BID) was found to be conserved at the C-terminus in a multiple sequence alignment in both strict and distant orthologs. The equal distribution of positively and negatively charged amino acid residues is found to be conserved throughout orthologs of the human C6orf222 protein.
Evolution
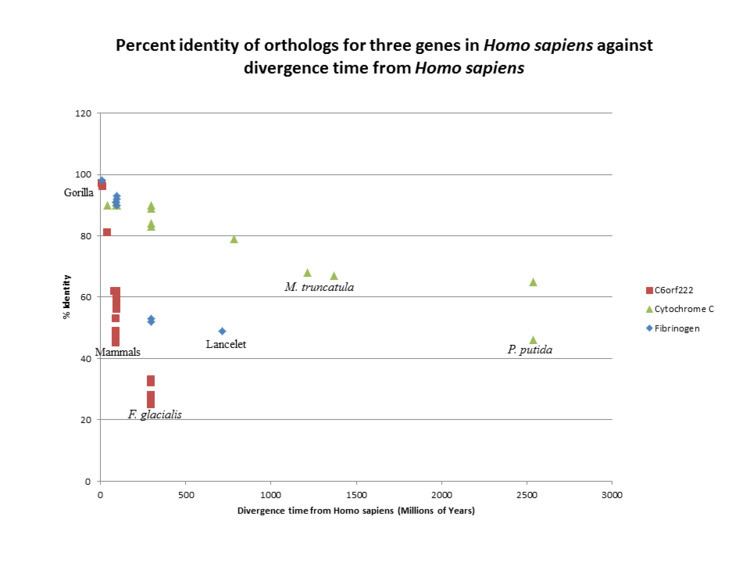
C6orf222 is a fast evolving gene, similar in nature to Fibrinogen alpha chain FGA. This evolutionary trend is in contrast to Cytochrome c, which has been shown to be a slow evolving gene.
