 | ||
Statistical shape analysis and statistical shape theory in computational anatomy (CA) is performed relative to templates, therefore it is a local theory of statistics on shape.Template estimation in computational anatomy from populations of observations is a fundamental operation ubiquitous to the discipline. Several methods for template estimation based on Bayesian probability and statistics in the random orbit model of CA have emerged for submanifolds and dense image volumes.
Contents
- The deformable template model of shapes and forms via diffeomorphic group actions
- Geodesic positioning via the Riemannian exponential
- The Bayes model of computational anatomy
- Surface templates for computational neuroanatomy and subcortical structures
- Surface estimation in cardiac computational anatomy
- MAP Estimation of volume templates from populations and the EM algorithm
- References
The deformable template model of shapes and forms via diffeomorphic group actions
Linear algebra is one of the central tools to modern engineering. Central to linear algebra is the notion of an orbit of vectors, with the matrices forming groups (matrices with inverses and identity) which act on the vectors. In linear algebra the equations describing the orbit elements the vectors are linear in the vectors being acted upon by the matrices. In computational anatomy the space of all shapes and forms is modeled as an orbit similar to the vectors in linear-algebra, however the groups do not act linear as the matrices do, and the shapes and forms are not additive. In computational anatomy addition is essentially replaced by the law of composition.
The central group acting CA defined on volumes in
Groups and group are familiar to the Engineering community with the universal popularization and standardization of linear algebra as a basic model
A popular group action is on scalar images,
For sub-manifolds
Several group actions in computational anatomy have been defined.
Geodesic positioning via the Riemannian exponential
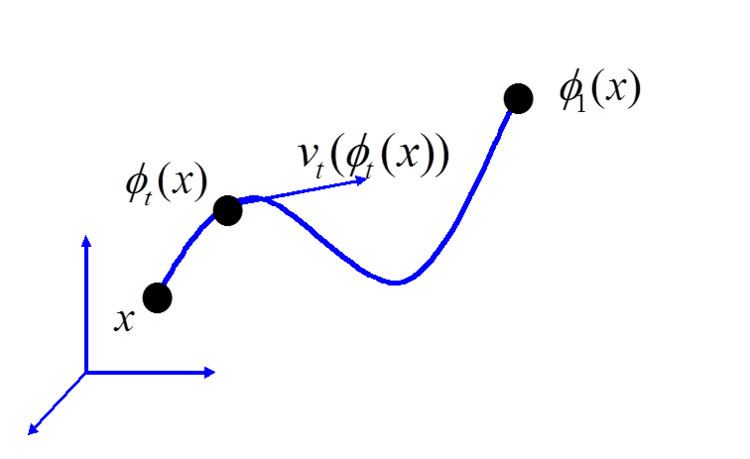
For the study of deformable shape in CA, a more general diffeomorphism group has been the group of choice, which is the infinite dimensional analogue. The high-dimensional diffeomorphism groups used in computational anatomy are generated via smooth flows
with
and the
Flows were first introduced for large deformations in image matching;
To ensure smooth flows of diffeomorphisms with inverse, the vector fields
The Bayes model of computational anatomy
The central statistical model of computational anatomy in the context of medical imaging is the source-channel model of Shannon theory; the source is the deformable template of images
Maximum a posteriori estimation (MAP) estimation is central to modern statistical theory. Parameters of interest
This requires computation of the conditional probabilities
Surface templates for computational neuroanatomy and subcortical structures
The study of sub-cortical neroanatomy has been the focus of many studies. Since the original publications by Csernansky and colleagues of hippocampal change in Schizophrenia, Alzheimer's disease, and Depression, many neuroanatomical shape statistical studies have now been completed using templates built from all of the subcortical structures for depression, Alzheimer's, Bipolar disorder, ADHD, autism, and Huntington's Disease. Templates were generated using Bayesian template estimation data back to Ma, Younes and Miller.
Shown in the accompanying Figure is an example of subcortical structure templates generated from T1-weighted magnetic resonance imagery by Tang et al. for the study of Alzheimer's disease in the ADNI population of subjects.
Surface estimation in cardiac computational anatomy
Numerous studies have now been done on cardiac hypertrophy and the role of the structural integraties in the functional mechanics of the heart. Siamak Ardekani has been working on populations of Cardiac anatomies reconstructing atlas coordinate systems from populations. The figure on the right shows the computational cardiac anatomy method being used to identify regional differences in radial thickness at end-systolic cardiac phase between patients with hypertrophic cardiomyopathy (left) and hypertensive heart disease (right). Color map that is placed on a common surface template (gray mesh) represents region ( basilar septal and the anterior epicardial wall) that has on average significantly larger radial thickness in patients with hypertrophic cardiomyopathy vs. hypertensive heart disease (reference below).
MAP Estimation of volume templates from populations and the EM algorithm
Generating templates empirically from populations is a fundamental operation ubiquitous to the discipline. Several methods based on Bayesian statistics have emerged for submanifolds and dense image volumes. For the dense image volume case, given the observable
In the Bayesian random orbit model of computational anatomy the observed MRI images
Ma's procedure for dense imagery takes an initial hypertemplate
The orbit-model is exploited by associating the unknown to be estimated flows to their log-coordinates
The EM algorithm takes as complete data the vector-field coordinates parameterizing the mapping,
