Symbol AT_hook InterPro IPR017956 SCOP 2eze | Pfam PF02178 SMART AT_hook SUPERFAMILY 2eze | |
 | ||
The AT-hook is a DNA-binding motif present in many proteins, including the high mobility group (HMG) proteins, DNA-binding proteins from plants and hBRG1 protein, a central ATPase of the human switching/sucrose non-fermenting (SWI/SNF) remodeling complex.
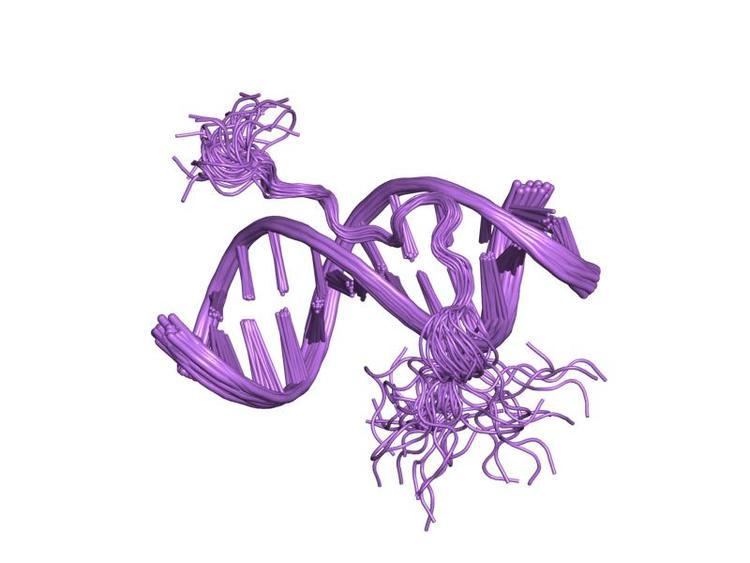
This motif consists of a conserved, palindromic, core sequence of proline-arginine-glycine-arginine-proline, although some AT-hooks contain only a single proline in the core sequence. AT-hooks also include a variable number of positively charged lysine and arginine residues on either side of the core sequence. The AT-hook binds to the minor groove of adenine-thymine (AT) rich DNA, hence the AT in the name. The rest of the name derives from a predicted asparagine/aspartate "hook" in the earliest AT-hooks reported in 1990. In 1997 structural studies using NMR determined that a DNA-bound AT-hook adopted a crescent or hook shape around the minor groove of a target DNA strand (pictured at right). HMGA proteins contain three AT-hooks, although some proteins contain as many as 30. The optimal binding sequences for AT-hook proteins are repeats of the form (ATAA)n or (TATT)n, although the optimal binding sequences for the core sequence of the AT-hook are AAAT and AATT.
