EC number 3.4.23.32 ExPASy NiceZyme view | CAS number 104781-89-7 | |
 | ||
Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B (EC 3.4.23.32, obsolete names include Scytalidium aspartic proteinase B, Ganoderma lucidum carboxyl proteinase, Ganoderma lucidum aspartic proteinase, Scytalidium lignicolum aspartic proteinase B, SLB) is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of the its molecular structure showed it to belong a novel group of proteases, glutamic protease.
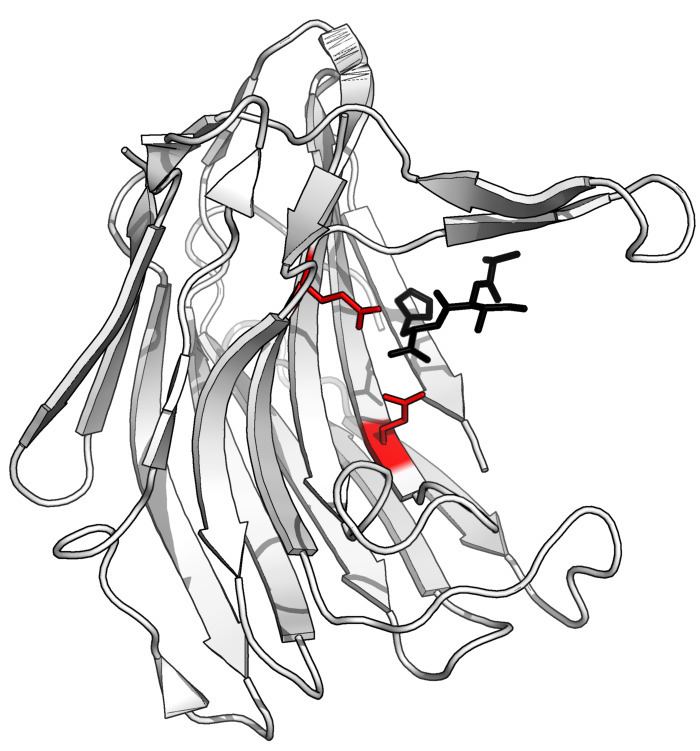
The protease has a unique structure and a novel catalytic dyad (E136 and Q53) in its active site. The active-site residues, glutamic acid (E) and glutamine (Q), was used to coin the name of the family of proteases, eqolisins, to which Scytalidoglutamic peptidase B belongs.
This enzyme catalyses the following chemical reaction
Hydrolysis of proteins with broad specificity, cleaving Phe24-Phe and Tyr26–Thr but not Leu15-Tyr and Phe25-Tyr in the B chain of insulin. It also cleaves the His6–Pro bond of angiotensin I, the ability to cleave a peptide bond with Pro in the P1′ position is unusual.This endopeptidase is isolated from Scytalidium lignicolum. It is an acid protease, and is most active at pH 2.0 when casein is used as substrate. Eqolosins prefer bulky amino acid residues at the P1 site and small amino acid residues at the P1′ site. The substrate specificity of scytalidoglutamic peptidase is unique, particularly in the substrate preferences at the P3 (basic amino acid), P1′ (small amino acid) and P3′ (basic) positions.
