 | ||
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is a technique for the amplification of RNA.
Contents
Within the last 10 years of its development, applications of the LAMP method in pathogenic microorganisms, genetically modified ingredients, tumor detection, and embryo sex identification have been widely used. This method was then improved by taking it a step further and combining it with a reverse transcription phase to allow for the detection of RNA. RT-LAMP is a one step nucleic acid amplification method that is used to diagnose infectious disease caused by bacteria or viruses. Although it has not been formally recognised by NAT, the method has been developed into many commercial kits that can be used for the identification of pathogens. The commonly used PCR method is able to generate millions of copies of the target strand. This process relies on thermal cycling, cycles of heating and cooling to facilitate the DNA replication. RT-LAMP does not require these cycles and is performed at a constant temperature between 60 and 65 °C. Similar to RT-PCR, RT-LAMP uses reverse transcriptase to synthesize complementary DNA (cDNA) from RNA sequences. This cDNA is then amplified using DNA polymerase, generating 10^9 copies per hour.
RT-LAMP is used in the detection of viruses. This method can be very effective in detecting viruses with an RNA genome (Group II, IV, and V based on the Baltimorevirus classification system).
Methodology
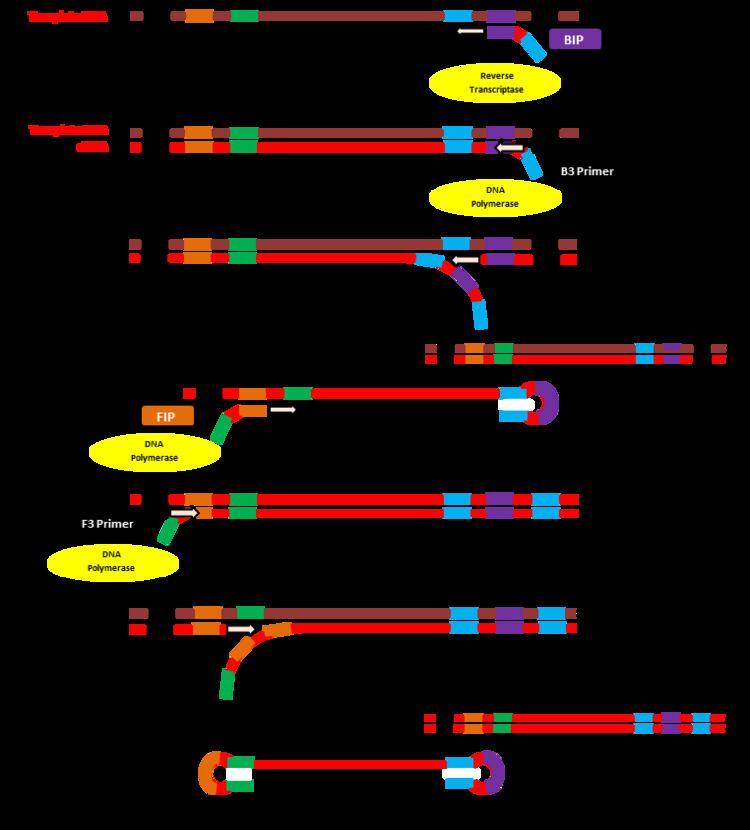
Four specially designed primers recognize distinct target sequences on the template strand. The primers bind only to these sequences which allows for high specificity. Out of the 4 primers involved, two of them are “inner primers” (FIP and BIP) which are designed to synthesize new DNA strands. The outer primers (F3 and B3) anneal to the template strand and also generate new DNA. These primers are accompanied by DNA polymerase which aids in strand displacement and releases the newly formed DNA strands.
The BIP primer, accompanied by reverse transcriptase, initiates the process by binding to a target sequence on the 3’ end of the RNA template and synthesizing a copy DNA strand. The B3 primer binds to this side of the template strand as well, and with the help of DNA polymerase simultaneously creates a new cDNA strand while displacing the previously made copy. The double stranded DNA containing the template strand is no longer needed.
The single stranded copy now loops at the 3’ end as it binds to itself. The FIP primer binds to the 5’ end of this single strand and accompanied by DNA polymerase, synthesizes a complementary strand. The F3 primer, with DNA polymerase, binds to this end and generates a new double stranded DNA molecule while displacing the previously made single strand.
This new single strand that has been released will act as the starting point for the LAMP cycling amplification. The DNA has a dumbbell-like structure as the ends fold in and self anneal. This structure becomes a stem-loop when the FIP or BIP primer once again initiates DNA synthesis at one of the target sequence locations. This cycle can be started from either the forward or backward side of the strand using the appropriate primer. Once this cycle has begun, the strand undergoes self-primed DNA synthesis during the elongation stage of the amplification process. This amplification takes place in only an hour, under isothermal conditions between 60-65 °C.
Advantages and disadvantages
This method is specifically advantageous because it can all be done quickly in one step.The sample is mixed with the primers, reverse transcriptase and DNA polymerase and the reaction takes place under a constant temperature. The required temperature is so low that the reaction can be completed using a simple hot water bath. There is no need for expensive thermocycling equipment that is necessary for other methods like PCR, which makes RT-LAMP very cost effective. In contrast with conventional PCR and real-time PCR assays, this method is much more efficient while still obtaining a high level of precision. This inexpensive and streamlined method can be more readily used in developing countries that do not have access to high tech laboratories. These areas are known for having a multitude of infectious diseases caused by various bacteria and viruses. The LAMP method is very useful for detection of these pathogens.
A disadvantage of this method is generating the sequence specific primers. For each LAMP assay, primers must be specifically designed to be compatible with the target DNA. This can be difficult which discourages researchers from using the LAMP method in their work. There is however, a free software called Primer Explorer, developed by Fujitsu in Japan, which can aid in the selection of these primers.
Applications
Viruses infect host cells with their specific genetic information, which the cell then replicates, causing the host to become diseased. In an effort to identify which certain virus is present in a host, RT-LAMP is used to test for the specific sequence of the virus, made possible by comparing the sequences against a large external database of references.
A primary example of the RT-LAMP was is an experiment to detect a new duck Tembusu-like, BYD virus, named after the region, Baiyangdian, where it was first isolated Because the symptoms of this virus were similar to Tembusu, an already identified disease, the nucleotide sequence of the complete genome of this virus was available in external resources. The known sequence was put into the online primer-designing softwear, LAMP Primer Explorer (http://primerexplorer.jp/e/), where the appropriate primers were designed and selected. With the selected primers, a RT-LAMP assay was done to amplify the RNA, with which the samples could then be visualized and confirmed under natural and UV light.
Another recent application of this method, was in a 2013 experiment to detect an Akabane virus using RT-LAMP. The experiment, done in China, isolated the virus from aborted calf fetuses, which is rarely successful but was able to be done because of RT-LAMP’s easy detection feature and high sensitivity. With the use of the Primer Explore V4 Software, and a sequence reference of the Akabane virus, the correct primers were developed and tested in an RT-LAMP assay. For specification purposes, the assay was also run against 4 other virus known to cause abortion in cattle. These comparative assays were unsuccessful due to the primers not binding to the template regions.
