 | ||
Proteasome accessory factor E ( PafE, also known as Bpa, Rv3780 ) is an ATP-independent proteasome activator of Mycobacterium tuberculosis that forms 12-fold symmetric rings and interacts with the 20S proteasome core particle through a conserved carboxyl-terminal motif to activate peptide and protein degradation.
Contents
Discovery
Mycobacterium tuberculosis (Mtb) is the etiologic agent of the disease tuberculosis, which kills approximately 1.4 million people each year. Mtb's arduous treatment regimen and the recent emergence of increasingly drug-resistant Mtb strains have given rise to an urgent need for the development of new anti-tubercular drugs. The mycobacterial proteasome, a large proteolytic complex is important for both Mtb virulence and resistance to the host's antimicrobial nitric oxide response, is a potential therapeutic target. Proteins can be targeted for proteasomal degradation in Mtb by the pupylation pathway, a process that is functionally analogous to eukaryotic ubiquitination. Pupylation centers around the small protein modifier Pup (prokaryotic ubiquitin-like protein), which is covalently ligated to a protein to target it for proteolysis. Despite advances in understanding of the Pup-proteasome system, no group has yet recapitulated robust degradation of a native substrate by the Mtb proteasome in vitro, implying that additional unknown factors are necessary for proteasomal activity. To search for such factors, researchers used a catalytically in active "proteasome trap" that stabilizes proteasomal interactions to identify novel proteasome-associated proteins, in 2014. PafE copurified with the proteasome trap in high abundance and co-occurs evolutionarily with the prokaryotic 20S core particle.
An M.tuberculosis PafE mutant accumulates misfolded proteins and is unable to quickly degrade HspR, resulting in the bacteria being unable to fully induce expression of its heat shock genes. As a result, the bacteria would be unable to cope with proteotoxic stress, leading to cell death.
Structure
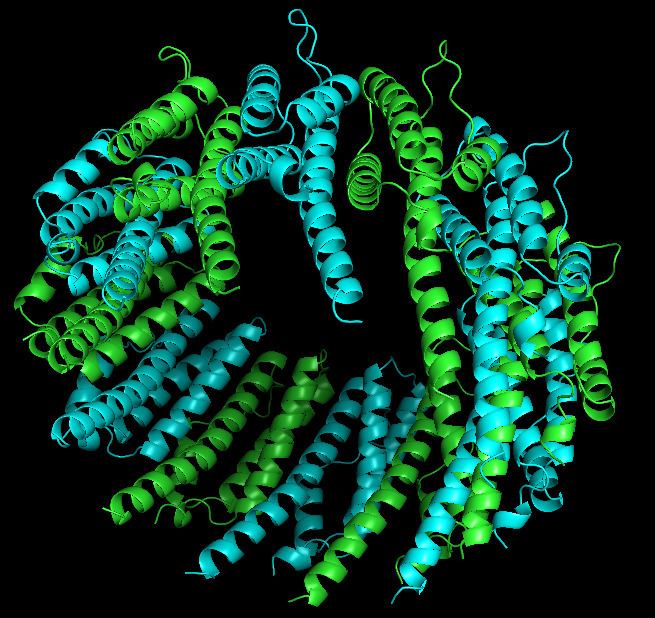
PafE forms a dodecameric ring structure with 12-fold symmetry. Each monomer is compose of 174 residues and forms four-helix bundle structure. The crystal structure revealed three unpredicted features of PafE. First, comparing with the previously known oligomeirc proeasome activators that have six- or sevenfold symmetry, PafE formed a dodecamer with 12-fold symmetry. The second feature is that PafE monomers formed a four-helix bundle structure similar to that of the eukaryotic PA26 and PA28 activator proteins, despite the absence of sequence homology between PafE and these proteins. Nevertheless, the length of the PafE alpha-helices on average is much shorter, and their packing is different from that of PA26 and PA28 in 11S activators. The third finding is that the orientation of a PafE subunit within the ring structure is different from that of PA26 or PA28: PafE helices (H) H1 and H4 face the interior channel but in Pa26 and PA28, H3 and H4 face inside.
Function
PafE interacts with the core proteasome alpha-subunit (PrcA) through its C-terminal hydrophobic-tyrosine-X motif (HbYX motif), as a switch to open gate of core proteasome. Interaction of PafE with the proteasome stimulates proteasomal peptidase and casein degradation activity, which suggests PafE could play a role in the removal of non-native or damaged proteins by influencing the conformation of the proteasome complex upon interaction. It can inhibit degradation of Pup-tagged substrates in vitro by competing with Mpa for association with the proteasome.
