Symbol Pup/Mpa PDB:3M9D Pfam structures PDBsum structure summary | Pfam PF05639 PDB RCSB PDB; PDBe; PDBj | |
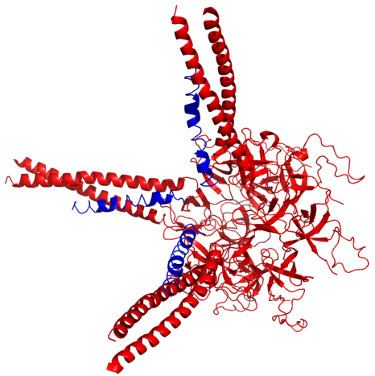 | ||
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote Mycobacterium tuberculosis. It serves the same function as ubiquitin, although the enzymology of ubiquitylation and pupylation is different. In contrast to the three-step reaction of ubiquitylation, pupylation requires two steps, therefore only two enzymes are involved in pupylation. Similar to ubiquitin, Pup attaches to specific lysine residues of substrate proteins by forming isopeptide bonds. It is then recognized by Mycobacterium proteasomal ATPase (Mpa) by a binding-induced folding mechanism that forms a unique alpha-helix. Mpa then delivers the Pup-substrate to the 20S proteasome by coupling of ATP hydrolysis for proteasomal degradation.
The discovery of Pup indicates that like eukaryotes, bacteria may use a small-protein modifier to control protein stability.
The X-ray crystal structure of the Pup/Mpa complex (PDB: 3M9D) was first determined by scientists Tao Wang and Huilin Li of the Brookhaven National Laboratory.
The Pup gene encodes a 64–amino acid protein with a molecular size of 6.944 kDa:
