 | ||
Similar Eobacteria, Caldisericum, Elusimicrobia | ||
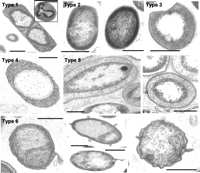
Poribacteria are a candidate phylum of bacteria originally identified in the microbiome of sea sponges (Porifera). Poribacteria were distinguished from other microorganisms associated with sea sponges by their distinctive morphology featuring a large membrane-bound cellular compartment that frequently contains DNA, a highly unusual feature for a prokaryote. Poribacteria are Gram-negative mixotrophs.
Contents
Genome
Single-cell genomics analysis of poribacteria reveals a genome with a lower size bound of 1.88 megabases and 1585 protein-coding genes, of which an unusually high 24% have no homology to known genes. Among the genes of identifiable homology, genetic infrastructure can be identified for aerobic metabolism, denitrification and urea uptake, and carbon fixation through the Wood-Ljungdahl pathway.
The poribacterial genome is also reported to contain an unusually high number of phyH-domain proteins, which are enzymes involved in oxidative reactions. The functional significance of this observation is not known.
Cell compartmentalization
Cell compartmentalization into distinct membrane-bound organelles is a universal and defining property of eukaryotes, but had not been not observed in prokaryotes other than the Planctomycetes before the identification of Poribacteria. The distinctive poribacterial compartments were originally identified using fluorescence in situ hybridization and electron microscopy and were found to frequently, but not always, contain DNA. Genomic evidence reveals the presence of proteins associated with compartmentalization, but not of membrane coat proteins.
Eukaryote-like proteins
Genomic analyses of poribacteria reveal several families of cell-surface repeat proteins that resemble those found in eukaryotes, and are infrequently found in prokaryotes. Examples include ankyrin and leucine-rich repeat domains, as well as tetratricopeptides. Unusual low-density lipoprotein receptor repeat proteins are also found, of unknown function. Most of these protein families are thought to be involved in surface interactions with the sponge host.
In addition, genetic infrastructure for sterol biosynthesis is observed in poribacterial genomes, otherwise found almost exclusively in eukaryotes and the planctomycete Gemmata obscuriglobus.
Ecological niche
Poribacteria are symbionts of sea sponges, among the most abundant microorganisms in the highly diverse microbiome of the sponge mesohyl. They have been found in a large variety of sponge species from diverse geographic origins. The distribution of microorganisms in the sponge microbiome can be vertically inherited, with adult sponges transmitting their distinctive microbial communities to offspring.
