Group Group I (dsDNA) | Rank Family | |
 | ||
Lower classifications | ||
Polyomaviridae is a family of viruses whose natural hosts are primarily mammals and birds. As of the most recent (2015) taxonomy release by the International Committee on Taxonomy of Viruses, there were 73 recognized species in this family contained within four genera, as well as three species that could not be assigned to a genus. Of these, 13 species are known to infect humans. Most of these viruses, such as BK virus and JC virus, are very common and typically asymptomatic in most human populations studied. However, some polyomaviruses are associated with human disease, particularly in immunocompromised individuals; BK virus is associated with nephropathy in renal transplant and non-renal solid organ transplant patients, JC virus with progressive multifocal leukoencephalopathy, and Merkel cell virus with Merkel cell cancer.
Contents
- Structure and genome
- Replication and life cycle
- Tumor antigens
- Agnoprotein
- Taxonomy
- Human polyomaviruses
- List of human polyomaviruses
- Clinical relevance
- SV40
- Diagnosis
- History
- References
Some members of the family are oncoviruses, meaning they can cause tumors; they often persist as latent infections in a host without causing disease, but may produce tumors in a host of a different species, or in individuals with ineffective immune systems. The family was first discovered due to its oncogenic properties; some members of the family, most prominently murine polyomavirus, have been extensively studied in the laboratory to understand the mechanism by which they induce carcinogenesis. The name polyoma refers to the viruses' ability to produce multiple (poly-) tumors (-oma).
Structure and genome
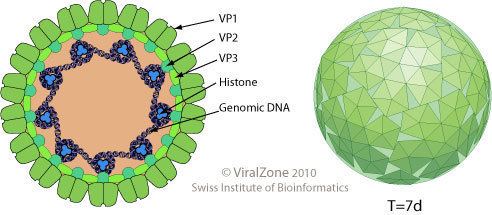
Polyomaviruses are unenveloped double-stranded DNA viruses with circular genomes of around 5000 base pairs. The genome is packaged in a viral capsid of about 40-50 nanometers in diameter, which is icosahedral in shape (T=7 symmetry). The capsid is composed of 72 pentameric capsomeres of a protein called VP1, which is capable of self-assembly into a closed icosahedron; each pentamer of VP1 is associated with one molecule of one of the other two capsid proteins, VP2 or VP3.
The genome of a typical polyomavirus codes for between 5 and 9 proteins, divided into two transcriptional regions called the early and late regions due to the time during infection in which they are transcribed. Each region is transcribed by the host cell's RNA polymerase II as a single pre-messenger RNA containing multiple genes. The early region usually codes for two proteins, the small and large tumor antigens, produced by alternative splicing. The late region contains the three capsid structural proteins VP1, VP2, and VP3, produced by alternative translational start sites. Additional genes and other variations on this theme are present in some viruses: for example, rodent polyomaviruses have a third protein called middle tumor antigen in the early region, which is extremely efficient at inducing cellular transformation; SV40 has an additional capsid protein VP4; some examples have an additional regulatory protein called agnoprotein expressed from the late region. The genome also contains a non-coding control or regulatory region containing the early and late regions' promoters, transcriptional start sites, and the origin of replication.
Replication and life cycle
The polyomavirus life cycle begins with entry into a host cell. Cellular receptors for polyomaviruses are sialic acid residues of glycans, commonly gangliosides. The attachment of polyomaviruses to host cells is mediated by the binding of VP1 to sialylated glycans on the cell surface. In some particular viruses, additional cell-surface interactions occur; for example, the JC virus is believed to require interaction with the 5HT2A receptor and the Merkel cell virus with heparan sulfate. However, in general virus-cell interactions are mediated by commonly occurring molecules on the cell surface, and therefore are likely not a major contributor to individual viruses' observed cell-type tropism. After binding to molecules on the cell surface, the virion is endocytosed and enters the endoplasmic reticulum - a behavior unique among known non-enveloped viruses - where the viral capsid structure is likely to be disrupted by action of host cell disulfide isomerase enzymes.
The details of transit to the nucleus are not clear and may vary among individual polyomaviruses. It has been frequently reported that an intact, albeit distorted, virion particle is released from the endoplasmic reticulum into the cell cytoplasm, where the genome is released from the capsid, possibly due to the low calcium concentration in the cytoplasm. Both expression of viral genes and replication of the viral genome occur in the nucleus using host cell machinery. The early genes - comprising at minimum the small tumor antigen (ST) and large tumor antigen (LT) - are expressed first, from a single alternatively spliced messenger RNA strand. These proteins serve to manipulate the host's cell cycle - dysregulating the transition from G1 phase to S phase, when the host cell's genome is replicated - because host cell DNA replication machinery is needed for viral genome replication. The precise mechanism of this dysregulation depends on the virus; for example, SV40 LT can directly bind host cell p53, but murine polyomavirus LT does not. LT induces DNA replication from the viral genome's non-coding control region (NCCR), after which expression of the early mRNA is reduced and expression of the late mRNA, which encodes the viral capsid proteins, begins. Several mechanisms have been described for regulating the transition from early to late gene expression, including the involvement of the LT protein in repressing the early promoter, the expression of un-terminated late mRNAs with extensions complementary to early mRNA, and the expression of regulatory microRNA.
Expression of the late genes results in accumulation of the viral capsid proteins in the host cell cytoplasm. Capsid components enter the nucleus in order to encapsidate new viral genomic DNA. New virions may be assembled in viral factories. The mechanism of viral release from the host cell varies among polyomaviruses; some express proteins that facilitate cell exit, such as the agnoprotein or VP4. In some cases high levels of encapsidated virus result in cell lysis, releasing the virions.
Tumor antigens
The large tumor antigen plays a key role in regulating the viral life cycle by binding to the viral origin of DNA replication where it promotes DNA synthesis. Also as the polyomavirus relies on the host cell machinery to replicate the host cell needs to be in s-phase for this to begin. Due to this, large T-antigen also modulates cellular signaling pathways to stimulate progression of the cell cycle by binding to a number of cellular control proteins. This is achieved by a two prong attack of inhibiting tumor suppressing genes p53 and members of the retinoblastoma (pRB) family, and stimulating cell growth pathways by binding cellular DNA, ATPase-helicase, DNA polymerase α association, and binding of transcription preinitiation complex factors. This abnormal stimulation of the cell cycle is a powerful force for oncogenic transformation.
The small tumor antigen protein is also able to activate several cellular pathways that stimulate cell proliferation. Polyomavirus small T antigens commonly target protein phosphatase 2A (PP2A), a key multisubunit regulator of multiple pathways including Akt, the mitogen-activated protein kinase (MAPK) pathway, and the stress-activated protein kinase (SAPK) pathway. Merkel cell polyomavirus small T antigen encodes a unique domain, called the LT-stabilization domain (LSD), that binds to and inhibits the FBXW7 E3 ligase regulating both cellular and viral oncoproteins. Unlike for SV40, the MCV small T antigen directly transforms rodent cells in vitro.
The middle tumor antigen is used in model organisms developed to study cancer, such as the MMTV-PyMT system where middle T is coupled to the MMTV promoter. There it functions as an oncogene, while the tissue where the tumor develops is determined by the MMTV promoter.
Agnoprotein
The agnoprotein is a small multifunctional phospho-protein found in the late coding part of the genome of some polyomaviruses, most notably BK virus, JC virus, and SV40. It is essential for proliferation in the viruses that express it and is thought to be involved in regulating the viral life cycle, particularly replication and viral exit from the host cell, but the exact mechanisms are unclear.
Taxonomy
The polyomaviruses are members of group I (dsDNA viruses). The classification of Polyomaviruses has been the subject of several proposed revisions as new members of the group are discovered. Formerly, polyomaviruses and papillomaviruses, which share many structural features but have very different genomic organizations, were classified together in the now-obsolete family Papovaviridae. (The name Papovaviridae derived from three abbreviations: Pa for Papillomavirus, Po for Polyomavirus, and Va for "vacuolating.") The polyomaviruses were divided into three major clades (that is, genetically-related groups): the SV40 clade, the avian clade, and the murine polyomavirus clade. A subsequent proposed reclassification by the International Committee on Taxonomy of Viruses (ICTV) recommended dividing the family of Polyomaviridae into three genera:
The current ICTV classification system recognises four genera and 76 species, of which three could not be assigned a genus. This system retains the distinction between avian and mammalian viruses, grouping the avian subset into the genus Gammapolyomavirus.
Additional viruses continue to be described. These include the sea otter polyomavirus 1 and Alpaca polyomavirus
Human polyomaviruses
Most polyomaviruses do not infect humans. Of the polyomaviruses cataloged as of 2016, a total of 13 were known with human hosts. Many human polyomaviruses are very common and are asymptomatic. However, some polyomaviruses are associated with human disease, particularly in immunocompromised individuals. MCV is highly divergent from the other human polyomaviruses and is most closely related to murine polyomavirus. Trichodysplasia spinulosa-associated polyomavirus (TSV) is distantly related to MCV. Two viruses—HPyV6 and HPyV7—are most closely related to KI and WU viruses, while HPyV9 is most closely related to the African green monkey-derived lymphotropic polyomavirus (LPV).
List of human polyomaviruses
The following 13 polyomaviruses with human hosts had been identified and had their genomes sequenced as of 2016:
The proposed Deltapolyomavirus genus contains only the four human viruses shown, with human polyomavirus 6 as the type species. The Alpha and Beta groups contain viruses that infect a variety of mammals. The Gamma group contains the avian viruses. Clinically significant disease associations are shown only where causality is expected.
Clinical relevance
All the polyomaviruses are highly common childhood and young adult infections. Most of these infections appear to cause little or no symptoms. These viruses are probably lifelong persistent among almost all adults. Diseases caused by human polyomavirus infections are most common among immunocompromised people; disease associations include BK virus with nephropathy in renal transplant and non-renal solid organ transplant patients, JC virus with progressive multifocal leukoencephalopathy, and Merkel cell virus (MCV) with Merkel cell cancer.
SV40
SV40 replicates in the kidneys of monkeys without causing disease, but can cause cancer in rodents under laboratory conditions. In the 1950s and early 1960s, well over 100 million people may have been exposed to SV40 due to previously undetected SV40 contamination of polio vaccine, prompting concern about the possibility that the virus might cause disease in humans. Although it has been reported as present in some human cancers, including brain tumors, bone tumors, mesotheliomas, and non-Hodgkin's lymphomas, accurate detection is often confounded by high levels of cross-reactivity for SV40 with widespread human polyomaviruses. Most virologists dismiss SV40 as a cause for human cancers.
Diagnosis
The diagnosis of polyomavirus almost always occurs after the primary infection as it is either asymptomatic or sub-clinical. Antibody assays are commonly used to detect presence of antibodies against individual viruses. Competition assays are frequently needed to distinguish among highly similar polyomaviruses.
In cases of progressive multifocal leucoencephalopathy (PML), a cross-reactive antibody to SV40 T antigen (commonly Pab419) is used to stain tissues directly for the presence of JC virus T antigen. PCR can be used on a biopsy of the tissue or cerebrospinal fluid to amplify the polyomavirus DNA. This allows not only the detection of polyomavirus but also which sub type it is.
There are three main diagnostic techniques used for the diagnosis of the reactivation of polyomavirus in polyomavirus nephropathy (PVN): urine cytology, quantification of the viral load in both urine and blood, and a renal biopsy. The reactivation of polyomavirus in the kidneys and urinary tract causes the shedding of infected cells, virions, and/or viral proteins in the urine. This allows urine cytology to examine these cells, which if there is polyomavirus inclusion of the nucleus, is diagnostic of infection. Also as the urine of an infected individual will contain virions and/or viral DNA, quanitation of the viral load can be done through PCR. This is also true for the blood.
Renal biopsy can also be used if the two methods just described are inconclusive or if the specific viral load for the renal tissue is desired. Similarly to the urine cytology, the renal cells are examined under light microscopy for polyomavirus inclusion of the nucleus, as well as cell lysis and viral partials in the extra cellular fluid. The viral load as before is also measure by PCR.
Tissue staining using a monoclonal antibody against MCV T antigen shows utility in differentiating Merkel cell carcinoma from other small, round cell tumors. Blood tests to detect MCV antibodies have been developed and show that infection with the virus is widespread although Merkel cell carcinoma patients have exceptionally higher antibody responses than asymptomatically infected persons.
History
Murine polyomavirus was the first polyomavirus discovered, having been reported by Ludwik Gross in 1953 as an extract of mouse leukemias capable of inducing parotid gland tumors. The causative agent was identified as a virus by Sarah Stewart and Bernice Eddy, after whom it was once called "SE polyoma". The term "polyoma" refers to the viruses' ability to produce multiple (poly-) tumors (-oma) under certain conditions. The name has been criticized as a "meatless linguistic sandwich" ("meatless" because both morphemes in "polyoma" are affixes) giving little insight into the viruses' biology; in fact, subsequent research has found that most polyomaviruses rarely cause clinically significant disease in their host organisms under natural conditions.
Dozens of polyomaviruses have been identified and sequenced as of 2016, infecting mainly birds and mammals. Two polyomaviruses are known to infect fish, the black sea bass and gilthead seabream. A total of thirteen polyomaviruses are known to infect humans.
