Symbol Cyclase_polyket InterPro IPR006765 PDB RCSB PDB; PDBe; PDBj | Pfam PF04673 Pfam structures PDBsum structure summary | |
 | ||
In molecular biology, the polyketide synthesis cyclase family of proteins includes a number of cyclases involved in polyketide synthesis in a number of actinobacterial species.
Aromatic polyketides are assembled by a type II (iterative) polyketide synthase in bacteria. Iterative type II polyketide synthases produce polyketide chains of variable but defined length from a specific starter unit and a number of extender units. They also specify the initial regiospecific folding and cyclisation pattern of nascent polyketides either through the action of a cyclase (CYC) subunit or through the combined action of site-specific ketoreductase and CYC subunits. Additional CYCs and other modifications may be necessary to produce linear aromatic polyketides.
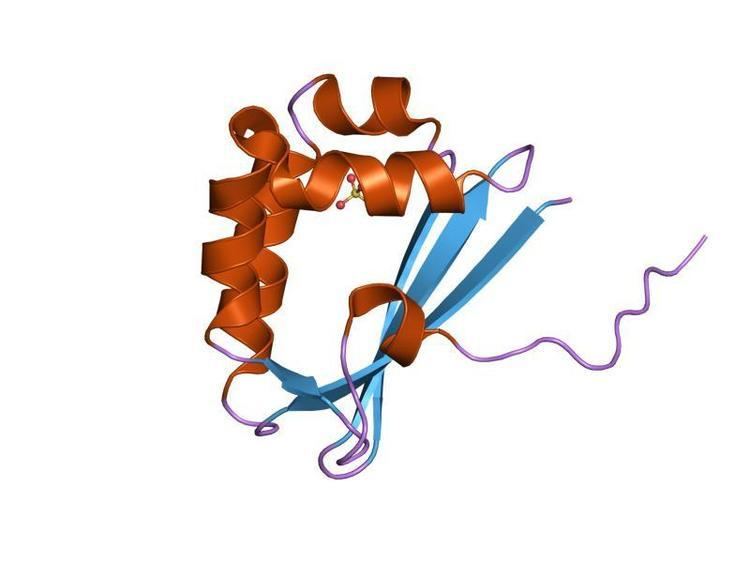
The Tetracenomycin polyketide synthesis protein, tcmI, from Streptomyces glaucescens catalyses an aromatic rearrangement in the biosynthetic pathaway of tetracenomycin C from Streptomyces coelicolor. The protein is a homodimer where each subunit forms a beta-alpha-beta fold belonging to the ferrodoxin fold superfamily. Four strands of antiparallel sheets and a layer of alpha helices create a cavity which was proposed to be the active site. This structure shows strong topological similarity to a polyketide monoxygenase from Streptomyces coelicolor which functions in the actinorhodin biosynthesic pathway. It was suggested, therefore, that this fold is well suited to serve as a framework for rearrangements and chemical modification of polyaromatic substrates.
