 | ||
Nuclear receptor coregulators are a class of transcription coregulators that have been shown to be involved in any aspect of signaling by any member of the nuclear receptor superfamily. A comprehensive database of nuclear receptor coregulators can be found at the Nuclear Receptor Signaling Atlas website.
Contents
Introduction
The ability of nuclear receptors to alternate between activation and repression in response to specific molecular cues, is now known to be attributable in large part to a diverse group of cellular factors, collectively termed coregulators and including coactivators and corepressors. The study of nuclear receptors owed a debt to decades of historical endocrinology and pathology, and prior to their discovery there was a wealth of empirical evidence that suggested their existence. Coregulators, in contrast, have been the subject of a rapid accumulation of functional and mechanistic data which is yet to be consolidated into an integrated picture of their biological functions. While this article refers to the historical terms "coactivator" and "corepressor" it should be noted that this distinction is less clear than was at first thought, and it is now known that cell type, cell signaling state and promoter identity can influence the direction of action of any given coregulator.
Coregulators are often incorrectly referred to as cofactors, which are small, non-protein molecules required by an enzyme for full activity, e.g. NAD+.
Coactivators
See also coactivators
As far back as the early 1970s, receptor-associated nonhistone proteins were known to support the function of nuclear receptors. In the early 1990s, some investigators such as Keith Yamamoto had suggested a role for non-DNA nuclear acceptor molecules. A biochemical strategy designed in Myles Brown’s laboratory provided the first direct evidence of ligand-dependent recruitment by nuclear receptors of ancillary molecules.
The yeast two-hybrid protein-protein interaction assay led to the identification of an array of receptor-interacting factors in David Moore’s laboratory and RIP140 repressive protein was discovered in Malcolm Parker’s laboratory.
The stage was now set for the cloning of the coactivators. The first authentic, common nuclear receptor coactivator was steroid receptor coactivator 1, or SRC-1, first cloned in Bert O’Malley’s laboratory. SRC-1 and two related proteins, GRIP-1, cloned first by Michael Stallcup, and ACTR/p/CIP, initially identified in Ron Evans and Geoff Rosenfeld’s lab, together make up the SRC family of coactivators. The SRC family is defined by the presence in the N-terminus of tandem PAS and beta-HLH motifs; a centrally-located domain which binds the coactivators CBP and p300; and a C-terminal region which mediates interaction with the CARM-1 coactivator. Malcolm Parker’s laboratory was the first to show that a recurring structural feature of many coactivators is an alpha-helical LXXLL motif (a contiguous sequence of 5 amino acids where L = leucine and X = any amino acid), or nuclear receptor box, present from a single to several copies in many coactivators, which is implicated in their ligand-dependent recruitment by the receptor AF-2. The SRC coactivator family, for example, has a conserved cluster of NR boxes located in the central region of each member of the family.
Coactivators can be categorized based upon their varied functional properties. To name a few, classes of coactivators include:
Corepressors
See also corepressors
Transcriptional repression by corepressors is in many ways conceptually comparable to the mediation of receptor transcriptional activation by coactivators, but has an opposite outcome. Recruitment of corepressors, generally occurring in the absence of ligand, depends on a critical conformation of the receptor AF-2 domain, as well as upon nuclear receptor box-like helical motifs in the corepressor. Moreover, corepressors themselves recruit ancillary enzyme activities which help to establish or maintain the repressive state at their target promoters.
Early cell transfection experiments had shown that discrete regions of certain receptors, such as thyroid hormone receptor, were sufficient to repress, or silence, reporter genes when fused to DNA-binding domains of heterologous transcription factors, suggesting that specific cellular factors – or corepressors - might bind to these regions and silence receptors in cells.
Again, using the yeast two-hybrid screen, two corepressors were isolated in rapid succession, nuclear receptor corepressor, or NCoR, in Geoff Rosenfeld’s laboratory, and silencing mediator of retinoid and thyroid receptors, or SMRT, by Ron Evans. Alignment of the two proteins indicated that they had a largely common domain structure, suggesting parallels in their mode of action. Mitch Lazar’s group has shown that inactive nuclear receptors recruit corepressors in part through amphipathic helical peptides called CoRNR boxes, which are similar to the coactivator nuclear receptor boxes.
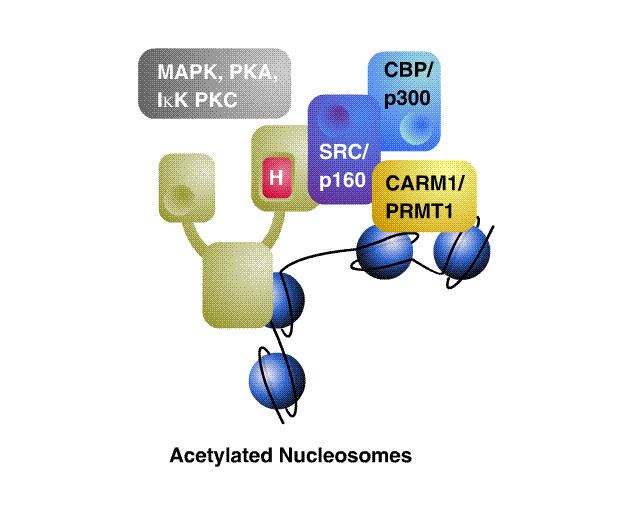
In addition to these structural analogies, corepressors and coactivators have common functional themes. The acetylation state of nucleosomes on a promoter is related to the rate of transcription of the gene. Histone acetylase coactivators increase the rate of acetylation, opening the nucleosome to transcription factors; histone deacetylases recruited by corepressors reverse this reaction, silencing transcription of the target gene. Other histone modifications have similar or opposite effects on transcription.
Biology of coregulators
The physiological role of SRC/p160s, CBP/p300 and other coactivators has been implied by knockout studies in mice of genes encoding these proteins. The effects of these deletions range from the profound effects on viability characteristic of TRAP220, CBP and p300, to the more subtle developmental and metabolic phenotypes associated with members of the SRC family. Using sequences from cloned coregulator genes, laboratories such as those led by Bert O’Malley (SRC-1), Bob Roeder (TRAP220), Geoff Rosenfeld (NCoR), and Pierre Chambon (GRIP1) were able to delete, or knockout these genes in mice. These studies showed that coactivators were required for physiological and developmental functions of steroid and thyroid hormones in living animals, and that corepressors too have crucial roles in the development of certain organs.
Regulation of coregulator function
A spectrum of post-translational modifications is known to regulate the functional relationships between nuclear receptors, their coregulator complexes, and their target gene networks. Targeted, reversible enzymatic modifications such as acetylation, methylation phosphorylation and terminal modifications such as ubiquitination have been shown to have a variety of effects on coregulator function. Coregulators may be viewed as control interfaces for integrating multiple afferent stimuli into an appropriate cellular response. One possible scenario is that differential phosphorylation of coactivators may direct their combinatorial recruitment into different transcriptional complexes at distinct promoters in specific cells.
General model
Coactivators exist in large, modular complexes in the cell, and are known to participate in many different protein-protein interactions. A current model is that the composition of these complexes can become fluid, mixing and matching subunits to tailor the specific needs of different receptors, ligands or promoters. While spatiotemporal aspects of nuclear receptor and coregulator action remain poorly defined, a broad composite model of nuclear receptor action invokes corepressors as critical mediators of nuclear receptor silencing. In turn, a variety of coactivators are implicated in transcriptional activation by nuclear receptors, including SWI/SNF chromatin remodeling machines, SRC/p160s and TRAP/DRIP. The model accommodates the ability of membrane G protein coupled signaling pathways and tyrosine receptor signaling to cross-talk with coactivator and corepressor functions at the transcriptional level.
Coregulators and human disease
With the well-documented role of nuclear receptor coregulators in a variety of molecular functions within the cell, it should come as no surprise that evidence implicates them in a wide variety of diseases states, including cancer, metabolic syndromes (obesity, diabetes) and heritable syndromes such as Rubinstein-Taybi Syndrome, Angelman Syndrome and Von Gierke's disease. A comprehensive review of the role of coregulators in human disease has been published, which shows that over 165 of the known coregulators have been implicated in human pathologies.
