 | ||
Nanopore sequencing is a method under development since 1995 for sequencing DNA (determining the order in which nucleotides occur on a strand of DNA).
Contents
- Background
- Principle of nanopore sequencing
- Alpha hemolysin
- MspA
- Fluorescence
- Electron tunneling
- Challenges
- Commercialization
- Reviews
- References
A nanopore is simply a small hole with an internal diameter of the order of 1 nanometer. Certain porous transmembrane cellular proteins act as nanopores, and nanopores have also been made by etching a somewhat larger hole (several tens of nanometers) in a piece of silicon, and then gradually filling it in using ion-beam sculpting methods which results in a much smaller diameter hole: the nanopore. Graphene is also being explored as a synthetic substrate for solid-state nanopores.
The theory behind nanopore sequencing is that when a nanopore is immersed in a conducting fluid and a potential (voltage) is applied across it, an electric current due to conduction of ions through the nanopore can be observed. The amount of current is very sensitive to the size and shape of the nanopore. If single nucleotides (bases), strands of DNA or other molecules pass through or near the nanopore, this can create a characteristic change in the magnitude of the current through the nanopore.
Background
DNA could be passed through the nanopore for various reasons. For example, electrophoresis might attract the DNA towards the nanopore, and it might eventually pass through it. Or, enzymes attached to the nanopore might guide DNA towards the nanopore. The scale of the nanopore means that the DNA may be forced through the hole as a long string, one base at a time, rather like threading through the eye of a needle. As it does so, each nucleotide on the DNA molecule may obstruct the nanopore to a different, characteristic degree. The amount of current which can pass through the nanopore at any given moment therefore varies depending on whether the nanopore is blocked by an A, a C, a G or a T or a section of DNA that includes more than one of these bases (kmer). The change in the current through the nanopore as the DNA molecule passes through the nanopore represents a direct reading of the DNA sequence.
Alternatively, a nanopore might be used to identify individual DNA bases as they pass through the nanopore in the correct order.
Using Nanopore sequencing, a single molecule of DNA can be sequenced directly using a nanopore, without the need for an intervening PCR amplification step or a chemical labelling step or the need for optical instrumentation to identify the chemical label. The versatility of the nanopore concept is underlined by the fact that it has also been proposed for detection of life on other planets, since it is not necessarily restricted to the detection of the genetic information carrier DNA, but in general can be applied to sequence chain-like genetic information carriers without knowing the exact structure of their building blocks. Publications on the method outline its use in rapid identification of viral pathogens, monitoring ebola, environmental monitoring, food safety monitoring, human genome sequencing, plant genome sequencing, monitoring of antibiotic resistance, haplotyping and other applications.
Principle of nanopore sequencing
In nanopore sequencing, the nanopore is embedded in a membrane to create a passage for the ionic solution. When a constant voltage is added over each side of the membrane, an ionic current through the passage is detected and drive ssDNA or ssRNA to pass through the pore. The polynucleotides are bounded with polymerase or helicase enzyme to control the movement speed of nucleotide. The narrowest region of nanopore is most sensitive. The nucleobase's mass and its associated electrical field will change the ionic conductivity of the sensitive region, resulting in a current level variation. The ionic current levels reveal the sequence of strand and can be measured by a sensitive ammeter.
Alpha hemolysin
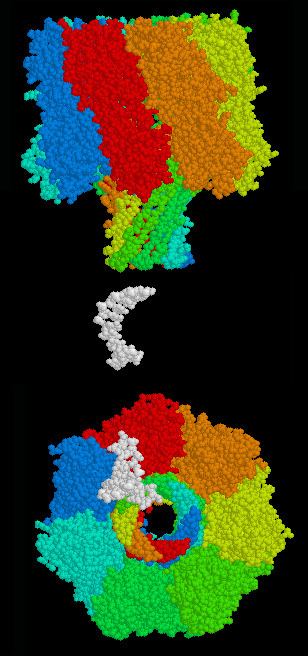
Alpha hemolysin (αHL), a nanopore from bacteria that causes lysis of red blood cells, has been studied for over 15 years. To this point, studies have shown that all four bases can be identified using ionic current measured across the αHL pore. The structure of αHL is advantageous to identify specific bases moving through the pore. The αHL pore is ~10 nm long, with two distinct 5 nm sections. The upper section consists of a larger, vestibule-like structure and the lower section consists of three possible recognition sites (R1, R2, R3), and is able to discriminate between each base.
Sequencing using αHL has been developed through basic study and structural mutations, moving towards the sequencing of very long reads. Protein mutation of αHL has improved the detection abilities of the pore. The next proposed step is to bind an exonuclease onto the αHL pore. The enzyme would periodically cleave single bases, enabling the pore to identify successive bases. Coupling an exonuclease to the biological pore would slow the translocation of the DNA through the pore, and increase the accuracy of data acquisition.
Notably, theorists have shown that sequencing via exonuclease enzymes as described here is not feasible. This is mainly due to diffusion related effects imposing a limit on the capture probability of each nucleotide as it is cleaved. This results in a significant probability that a nucleotide is either not captured before it diffuses into the bulk or captured out of order, and therefore is not properly sequenced by the nanopore, leading to insertion and deletion errors. Therefore, major changes are needed to this method before it can be considered a viable strategy.
A recent study has pointed to the ability of αHL to detect nucleotides at two separate sites in the lower half of the pore. The R1 and R2 sites enable each base to be monitored twice as it moves through the pore, creating 16 different measurable ionic current values instead of 4. This method improves upon the single read through the nanopore by doubling the sites that the sequence is read per nanopore.
MspA
Mycobacterium smegmatis porin A (MspA) is the second biological nanopore currently being investigated for DNA sequencing. The MspA pore has been identified as a potential improvement over αHL due to a more favorable structure. The pore is described as a goblet with a thick rim and a diameter of 1.2 nm at the bottom of the pore. A natural MspA, while favorable for DNA sequencing because of shape and diameter, has a negative core that prohibited single stranded DNA(ssDNA) translocation. The natural nanopore was modified to improve translocation by replacing three negatively charged aspartic acids with neutral asparagines.
The electric current detection of nucleotides across the membrane has been shown to be tenfold more specific than αHL for identifying bases. Utilizing this improved specificity, a group at the University of Washington has proposed using double stranded DNA (dsDNA) between each single stranded molecule to hold the base in the reading section of the pore. The dsDNA would halt the base in the correct section of the pore and enable identification of the nucleotide. A recent grant has been awarded to a collaboration from UC Santa Cruz, the University of Washington, and Northeastern University to improve the base recognition of MspA using phi29 polymerase in conjunction with the pore.
Fluorescence
An effective technique to determine a DNA sequence has been developed using solid state nanopores and fluorescence. This fluorescence sequencing method converts each base into a characteristic representation of multiple nucleotides which bind to a fluorescent probe strand-forming dsDNA. With the two color system proposed, each base is identified by two separate fluorescences, and will therefore be converted into two specific sequences. Probes consist of a fluorophore and quencher at the start and end of each sequence, respectively. Each fluorophore will be extinguished by the quencher at the end of the preceding sequence. When the dsDNA is translocating through a solid state nanopore, the probe strand will be stripped off, and the upstream fluorophore will fluoresce.
This sequencing method has a capacity of 50-250 bases per second per pore, and a four color fluorophore system (each base could be converted to one sequence instead of two), will sequence over 500 bases per second. Advantages of this method are based on the clear sequencing readout—using a camera instead of noisy current methods. However, the method does require sample preparation to convert each base into an expanded binary code before sequencing. Instead of one base being identified as it translocates through the pore, ~12 bases are required to find the sequence of one base.
Electron tunneling
Measurement of electron tunneling through bases as ssDNA translocates through the nanopore is an improved solid state nanopore sequencing method. Most research has focused on proving bases could be determined using electron tunneling. These studies were conducted using a scanning probe microscope as the sensing electrode, and have proved that bases can be identified by specific tunneling currents. After the proof of principle research, a functional system must be created to couple the solid state pore and sensing devices.
Researchers at the Harvard Nanopore group have engineered solid state pores with single walled carbon nanotubes across the diameter of the pore. Arrays of pores are created and chemical vapor deposition is used to create nanotubes that grow across the array. Once a nanotube has grown across a pore, the diameter of the pore is adjusted to the desired size. Successful creation of a nanotube coupled with a pore is an important step towards identifying bases as the ssDNA translocates through the solid state pore.
Another method is the use of nanoelectrodes on either side of a pore. The electrodes are specifically created to enable a solid state nanopore's formation between the two electrodes. This technology could be used to not only sense the bases but to help control base translocation speed and orientation.
Challenges
One challenge for the 'strand sequencing' method was in refining the method to improve its resolution to be able to detect single bases. In the early papers methods, a nucleotide needed to be repeated in a sequence about 100 times successively in order to produce a measurable characteristic change. This low resolution is because the DNA strand moves rapidly at the rate of 1 to 5μs per base through the nanopore. This makes recording difficult and prone to background noise, failing in obtaining single-nucleotide resolution. The problem is being tackled by either improving the recording technology or by controlling the speed of DNA strand by various protein engineering strategies and Oxford Nanopore employs a 'kmer approach', analyzing more than one base at any one time so that stretches of DNA are subject to repeat interrogation as the strand moves through the nanopore one base at a time. Various techniques including algorithmic have been used to improve the performance of the MinION technology since it was first made available to users. More recently effects of single bases due to secondary structure or released mononucleotides have been shown.
Professor Hagan Bayley proposed in 2010 that creating two recognition sites within an alpha hemolysin pore may confer advantages in base recognition.
One challenge for the 'exonuclease approach', where a processive enzyme feeds individual bases, in the correct order, into the nanopore, is to integrate the exonuclease and the nanopore detection systems. In particular, the problem is that when an exonuclease hydrolyzes the phosphodiester bonds between nucleotides in DNA, the subsequently released nucleotide is not necessarily guaranteed to directly move in to, say, a nearby alpha-hemolysin nanopore. One idea is to attach the exonuclease to the nanopore, perhaps through biotinylation to the beta barrel hemolsyin. The central pore of the protein may be lined with charged residues arranged so that the positive and negative charges appear on opposite sides of the pore. However, this mechanism is primarily discriminatory and does not constitute a mechanism to guide nucleotides down some particular path.
Commercialization
Agilent Laboratories was the first to license and develop nanopores but does not have any current disclosed research in the area.
