Species Human Entrez 4609 | Human Mouse Ensembl ENSG00000136997 | |
 | ||
Aliases MYC, MRTL, MYCC, bHLHe39, c-Myc, v-myc avian myelocytomatosis viral oncogene homolog External IDs MGI: 97250 HomoloGene: 31092 GeneCards: MYC | ||
Myc (c-Myc) is a regulator gene that codes for a transcription factor. The protein encoded by this gene is a multifunctional, nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis and cellular transformation.
Contents
A mutated version of Myc is found in many cancers, which causes Myc to be constitutively (persistently) expressed. This leads to the unregulated expression of many genes, some of which are involved in cell proliferation, and results in the formation of cancer. A common human translocation involving Myc is critical to the development of most cases of Burkitt lymphoma. Malfunctions in Myc have also been found in carcinoma of the cervix, colon, breast, lung and stomach. Myc is thus viewed as a promising target for anti-cancer drugs.
In the human genome, Myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes through binding on enhancer box sequences (E-boxes) and recruiting histone acetyltransferases (HATs). This means that in addition to its role as a classical transcription factor, Myc also functions to regulate global chromatin structure by regulating histone acetylation both in gene-rich regions and at sites far from any known gene.
Discovery
Myc gene was first discovered in Burkitt lymphoma patients. In Burkitt lymphoma, cancer cells show chromosomal translocations, in which chromosome 8 is frequently involved. Cloning the break-point of the fusion chromosomes revealed a gene that was similar to myelocytomatosis viral oncogene (v-Myc). Thus, the newfound cellular gene was named c-Myc.
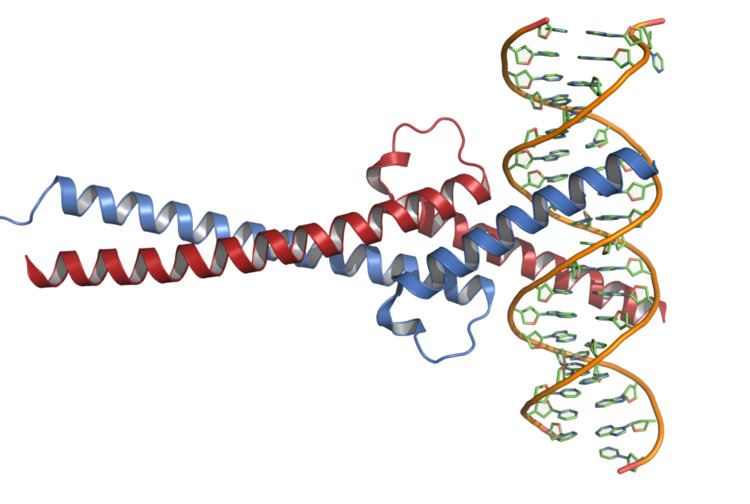
Structure
Myc protein belongs to Myc family of transcription factors, which also includes N-Myc and L-Myc genes. Myc family of transcription factors contain a bHLH (basic helix-loop-helix) structural and LZ (leucine zipper) motives. Through its bHLH DNA-binding motif, Myc interacts with DNA, while the leucine zipper TF-binding motif allows the dimerization with its partner Max, another bHLH transcription factor.
Myc mRNA contains an IRES (internal ribosome entry site) that allows the RNA to be translated into protein when 5' cap-dependent translation is inhibited, such as during viral infection.
Function
Myc protein is a transcription factor that activates expression of many genes through binding enhancer box sequences (E-boxes) and recruiting histone acetyltransferases (HATs). It can also act as a transcriptional repressor. By binding Miz-1 transcription factor and displacing the p300 co-activator, it inhibits expression of Miz-1 target genes. In addition, myc has a direct role in the control of DNA replication.
Myc is activated upon various mitogenic signals such as serum stimulation or by Wnt, Shh and EGF (via the MAPK/ERK pathway). By modifying the expression of its target genes, Myc activation results in numerous biological effects. The first to be discovered was its capability to drive cell proliferation (upregulates cyclins, downregulates p21), but it also plays a very important role in regulating cell growth (upregulates ribosomal RNA and proteins), apoptosis (downregulates Bcl-2), differentiation, and stem cell self-renewal. Myc is a very strong proto-oncogene and it is very often found to be upregulated in many types of cancers. Myc overexpression stimulates gene amplification, presumably through DNA over-replication.
There have been several studies that have clearly indicated Myc's role in cell competition.
A major effect of Myc is B cell proliferation.
c-Myc induces MTDH(AEG-1) gene expression and in turn itself requires AEG-1 oncogene for its expression.
Myc-nick
Myc-nick is a cytoplasmic form of Myc produced by a partial proteolytic cleavage of full-length c-Myc and N-Myc. Myc cleavage is mediated by the calpain family of calcium-dependent cytosolic proteases.
The cleavage of Myc by calpains is a constitutive process but is enhanced under conditions that require rapid downregulation of Myc levels, such as during terminal differentiation. Upon cleavage, the C-terminus of Myc (containing the DNA binding domain) is degraded, while Myc-nick, the N-terminal segment 298-residue segment remains in the cytoplasm. Myc-nick contains binding domains for histone acetyltransferases and for ubiquitin ligases.
The functions of Myc-nick are currently under investigation, but this new Myc family member was found to regulate cell morphology, at least in part, by interacting with acetyl transferases to promote the acetylation of α-tubulin. Ectopic expression of Myc-nick accelerates the differentiation of committed myoblasts into muscle cells.
Clinical significance
Except for early response genes, Myc universally upregulates gene expression. Furthermore, the upregulation is nonlinear. Genes whose expression is already significantly upregulated in the absence of Myc are strongly boosted in the presence of Myc, whereas genes whose expression is low in the absence Myc get only a small boost when Myc is present.
Inactivation of SUMO-activating enzyme (SAE1 / SAE2) in the presence of Myc hyperactivation results in mitotic catastrophe and cell death in cancer cells. Hence inhibitors of SUMOylation may be a possible treatment for cancer.
Amplification of the MYC gene was found in a significant number of epithelial ovarian cancer cases. In TCGA datasets, the amplification of Myc occurs in several cancer types, including breast, colorectal, pancreatic, gastric, and uterine cancers.
In the experimental transformation process of normal cells into cancer cells, the MYC gene can cooperate with the RAS gene.
Expression of Myc is highly dependent on BRD4 function in some cancers. BET inhibitors have been used to successfully block Myc function in pre-clinical cancer models and are currently being evaluated in clinical trials.
Animal Models
During the discovery of Myc gene, it was realized that chromosomes that reciprocally translocate to chromosome 8 contained immunoglobulin genes at the break-point. Enhancers that normally drive expression of immunoglobin genes now lead to overexpression of Myc proto-oncogene in lymphoma cells. To study the mechanism of tumorigenesis in Burkitt lymphoma by mimicking expression pattern of Myc in these cancer cells, transgenic mouse models were developed. Myc gene placed under the control of IgM heavy chain enhancer in transgenic mice gives rise to mainly lymphomas. Later on, in order to study effects of Myc in other types of cancer, transgenic mice that overexpress Myc in different tissues (liver, breast) were also made. In all these mouse models overexpression of Myc causes tumorigenesis, illustrating the potency of Myc oncogene. In a study with mice, reduced expression of Myc was shown to induce longevity, with significantly extended median and maximum lifespans in both sexes and a reduced mortality rate across all ages, better health, cancer progression was slower, better metabolism and they had smaller bodies. Also, Less TOR, AKT, S6K and other changes in energy and metabolic pathways (such as AMPK, more oxygen consumption, more body movements, etc). The study by John M. Sedivy and others used Cre-Loxp -recombinase to knockout one copy of Myc and this resulted in a "Haplo-insufficient" genotype noted as Myc+/-. The phenotypes seen oppose the effects of normal aging and are shared with many other long-lived mouse models such as CR (calorie restriction) ames dwarf, rapamycin, metformin and resveratrol. One study found that Myc and p53 genes were key to the survival of Chronic Myeloid Leukaemia (CML) cells. Targeting Myc and p53 proteins with drugs gave positive results on mice with CML.
Interactions
Myc has been shown to interact with:
