 | ||
The medcouple is a robust statistic that measures the skewness of a univariate distribution. It is defined as a scaled median difference of the left and right half of a distribution. Its robustness makes it suitable for identifying outliers in adjusted boxplots. Ordinary boxplots do not fare well with skew distributions, since they label the longer unsymmetrical tails as outliers. Using the medcouple, the whiskers of a boxplot can be adjusted for skew distributions and thus have a more accurate identification of outliers for non-symmetrical distributions.
Contents
- Definition
- Properties of the medcouple
- The medcouple kernel
- Robustness
- Values
- Algorithms for computing the medcouple
- Nave algorithm
- Fast algorithm
- Comparing a value against the kernel matrix
- Weighted median of row medians
- Kth pair algorithm
- Softwaresource code
- References
As a kind of order statistic, the medcouple belongs to the class of incomplete generalised L-statistics. Like the ordinary median or mean, the medcouple is a nonparametric statistic, thus it can be computed for any distribution.
Definition
In order to harmonise with zero-based indexing in many programming languages, we will index from zero in all that follows.
Let
of sizes
where
The medcouple is then the median of the set
In other words, we split the distribution into all values greater or equal to the median and all values less than or equal to the median. We define a kernel function whose first variable is over the
Since the medcouple is not a median applied to all
Properties of the medcouple
The medcouple has a number of desirable properties. A few of them are directly inherited from the kernel function.
The medcouple kernel
We make the following observations about the kernel function
- The kernel function is location-invariant. If we add or subtract any value to each element of the sample
X , the corresponding values of the kernel function do not change. - The kernel function is scale-invariant. Equally scaling all elements of the sample
X does not alter the values of the kernel function.
These properties are in turn inherited by the medcouple. Thus, the medcouple is independent of the mean and standard deviation of a distribution, a desirable property for measuring skewness. For ease of computation, these properties enable us to define the two sets
where
For
Using the recentred and rescaled set
- The kernel function is between -1 and 1, that is,
| h ( z i + , z j − ) | ≤ 1 . This follows from the reverse triangle inequality| a | − | b | ≤ | a − b | witha = z i + b = z j − z i + ≥ 0 ≥ z j − - The medcouple kernel
h ( z i + , z j − ) is non-decreasing in each variable. This can be verified by the partial derivatives∂ h ∂ z i + ∂ h ∂ z j − z i + ≥ 0 ≥ z j −
With properties 1, 2, and 4, we can thus define the following matrix,
If we sort the sets
The medcouple is then the median of this matrix with sorted rows and sorted columns. The fact that the rows and columns are sorted allows the implementation of a fast algorithm for computing the medcouple.
Robustness
The breakdown point is the number of values that a statistic can resist before it becomes meaningless, i.e. the number of arbitrarily large outliers that the data set
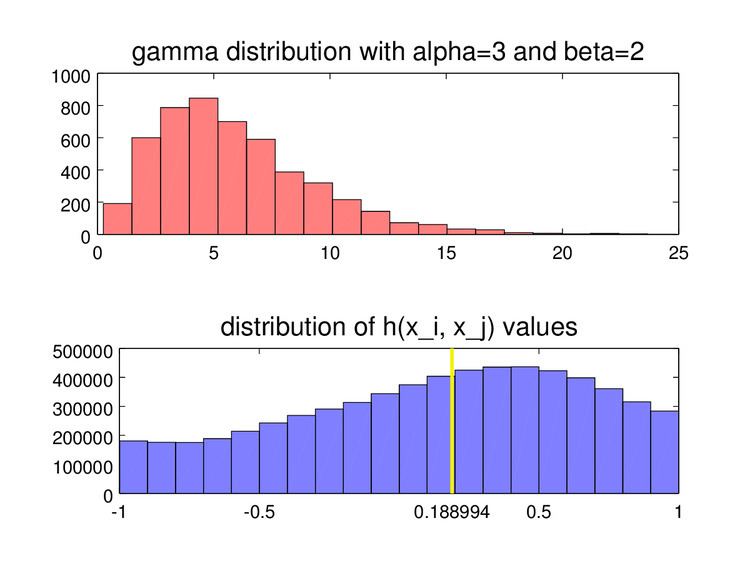
Values
Like all measures of skewness, the medcouple is positive for distributions that are skewed to the right, negative for distributions skewed to the left, and zero for symmetrical distributions. In addition, the values of the medcouple are bounded by 1 in absolute value.
Algorithms for computing the medcouple
Before presenting medcouple algorithms, we recall that there exist
Naïve algorithm
The naïve algorithm for computing the medcouple is slow. It proceeds in two steps. First, it constructs the medcouple matrix
More concretely, the naïve algorithm proceeds as follows. Recall that we are using zero-based indexing.
function naïve_medcouple(vector X): // X is a vector of size n. //Sorting in decreasing order can be done in-place in O(n log n) time sort_decreasing(X) xm := median(X) xscale := 2*max(X) // define the upper and lower centred and rescaled vectors // they inherit X's own decreasing sorting Zplus := [(x - xm)/xscale | x in X such that x >= xm] Zminus := [(x - xm)/xscale | x in X such that x <= xm] p := size(Zplus) q := size(Zminus) // define the kernel function closing over Zplus and Zminus function h(i,j): a := Zplus[i] b := Zminus[j] if a == b: return signum(p - 1 - i - j) else: return (a + b)/(a - b) endif endfunction // O(n^2) operations necessary to form this vector H := [h(i,j) | i in [0, 1, ..., p - 1] and j in [0, 1, ..., q - 1]] return median(H) endfunctionThe final call to median on a vector of size
Fast algorithm
The fast algorithm outperforms the naïve algorithm by exploiting the sorted nature of the medcouple matrix
The first stage of the fast algorithm proceeds as the naïve algorithm. We first compute the necessary ingredients for the kernel matrix,
Comparing a value against the kernel matrix
First, we note that we can compare any
This greater_h function is traversing the kernel matrix from the bottom left to the top right, and returns a vector
Conceptually, the resulting
The symmetric algorithm for computing the values of
This lower boundary can be visualised like so, where the blue entries are smaller than
For each
We also have that the sums
give, respectively, the number of elements of
Weighted median of row medians
The second observation is that we can use the sorted matrix structure to instantly compare any element to at least half of the entries in the matrix. For example, the median of the row medians across the entire matrix is less than the upper left quadrant in red, but greater than the lower right quadrant in blue:
More generally, using the boundaries given by the
The yellow entries indicate the median of each row. If we mentally re-arrange the rows so that the medians align and ignore the discarded entries outside the boundaries,
we can select a weighted median of these medians, each entry weighted by the number of remaining entries on this row. This ensures that we can discard at least 1/4 of all remaining values no matter if we have to discard the larger values in red or the smaller values in blue:
Each row median can be computed in
Kth pair algorithm
Putting together these two observations, the fast medcouple algorithm proceeds broadly as follows.
- Compute the necessary ingredients for the medcouple kernel function
h ( i , j ) withp sorted rows andq sorted columns. - At each iteration, approximate the medcouple with the weighted median of the row medians.
- Compare this tentative guess to the entire matrix obtaining right and left boundary vectors
P andQ respectively. The sum of these vectors also gives us the rank of this tentative medcouple.- If the rank of the tentative medcouple is exactly
p q / 2 , then stop. We have found the medcouple. - Otherwise, discard the entries greater than or less than the tentative guess by picking either
P orQ as the new right or left boundary, depending on which side the element of rankp q / 2 is in. This step always discards at least 1/4 of all remaining entries.
- If the rank of the tentative medcouple is exactly
- Once the number of candidate medcouples between the right and left boundaries is less than or equal to
p , perform a rank selection amongst the remaining entries, such that the rank within this smaller candidate set corresponds to thep q / 2 rank of the medcouple within the whole matrix.
The initial sorting in order to form the
Let us restate the fast algorithm in more detail.
function medcouple(vector X): // X is a vector of size n // compute initial ingredients as for the naïve medcouple sort_decreasing(X) xm := median(X) xscale := 2*max(X) Zplus := [(x - xm)/xscale | x in X such that x >= xm] Zminus := [(x - xm)/xscale | x in X such that x <= xm] p := size(Zplus) q := size(Zminus) function h(i,j): a := Zplus[i] b := Zminus[j] if a == b: return signum(p - 1 - i - j) else: return (a + b)/(a - b) endif endfunction // begin Kth pair algorithm (Johnson & Mizoguchi) // the initial left and right boundaries, two vectors of size p L := [0, 0, ..., 0] R := [q - 1, q - 1, ..., q - 1] // number of entries to the left of the left boundary Ltotal := 0 // number of entries to the left of the right boundary Rtotal := p*q // since we are indexing from zero, the medcouple index is one // less than its rank medcouple_index := floor(Rtotal/2) // iterate while the number of entries between the boundaries is // greater than the number of rows in the matrix while Rtotal - Ltotal > p: // compute row medians and their associated weights, but skip // any rows that are already empty middle_idx := [i | i in [0, 1, ..., p - 1] such that L[i] <= R[i]] row_medians := [h(i, floor((L[i] + R[i])/2) | i in middle_idx] weights := [R[i] - L[i] + 1 | i in middle_idx] WM := weighted median(row_medians, weights) // new tentative right and left boundaries P := greater_h(h, p, q, WM) Q := less_h(h, p, q, WM) Ptotal := sum(P) + size(P) Qtotal := sum(Q) // determine which entries to discard, or if we've found the medcouple if medcouple_index <= Ptotal - 1: R := P Rtotal := Ptotal else: if medcouple_index > Qtotal - 1: L := Q Ltotal := Qtotal else: // found the medcouple, rank of the weighted median equals medcouple index return WM endif endif endwhile // did not find the medcouple, but there are very few tentative entries remaining remaining := [h(i,j) | i in [0, 1, ..., p - 1], j in [L[i], L[i] + 1, ..., R[i]] such that L[i] <= R[i] ] // select the medcouple by rank amongst the remaining entries medcouple := select_nth(remaining, medcouple_index - Ltotal) return medcouple endfunctionIn real-world use, the algorithm also needs to account for errors arising from finite-precision floating point arithmetic. For example, the comparisons for the medcouple kernel function should be done within machine epsilon, as well as the order comparisons in the greater_h and less_h functions.
