EC number 3.2.1.106 ExPASy NiceZyme view | CAS number 78413-07-7 | |
 | ||
Mannosyl-oligosaccharide glucosidase (MOGS) (EC 3.2.1.106, processing alpha-glucosidase I, Glc3Man9NAc2 oligosaccharide glucosidase, trimming glucosidase I, GCS1) is an enzyme with systematic name mannosyl-oligosaccharide glucohydrolase. MOGS is a transmembrane protein found in the membrane of the endoplasmic reticulum of eukaryotic cells. Biologically, it functions within the N-glycosylation pathway.
Contents
Enzyme mechanism
MOGS is a glycoside hydrolase enzyme, belonging to Family 63 as classified within the Carbohydrate-Active Enzyme database.
This enzyme catalyses the following chemical reaction:
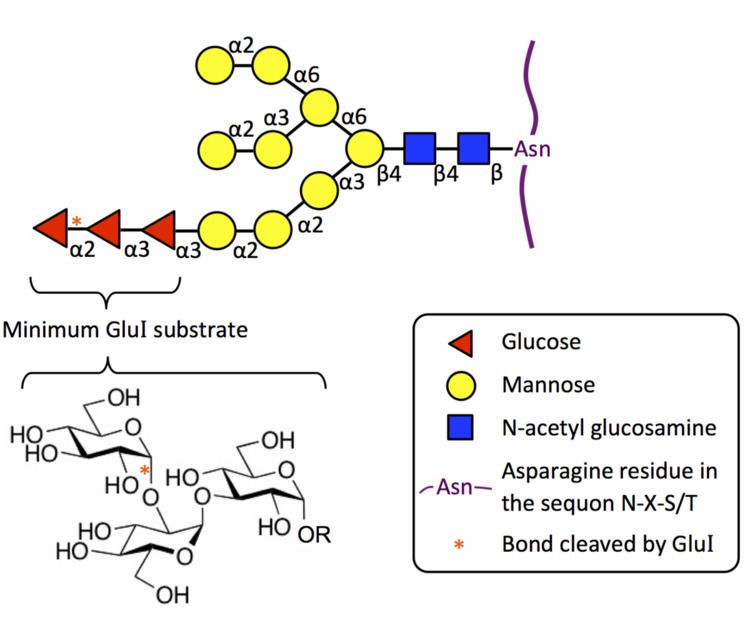
Exohydrolysis of the non-reducing terminal glucose residue in the mannosyl-oligosaccharide glycan Glc3Man9GlcNAc2This reaction is the first trimming step in the N-glycosylation pathway. Prior to this, the glycan was co-translationally attached to a nascent protein by the oligosaccharyltransferase complex. MOGS removes the terminal glucose residue, leaving the glycoprotein linked to Glc2Man9GlcNAc2, which can then serve as a substrate for glucosidase II.
Substrate Specificity
MOGS is highly specific to the oligosaccharide in its biological substrate in the N-glycosylation pathway. Eukaryotic MOGS does not cleave simple substrates such as p-nitrophenyl glucose, and it also shows no activity to the α(1→3) linkage present at the terminus of Glc1-2Man9GlcNAc2. Furthermore, the minimum substrate is the glucotriose molecule (Glc-α(1→2)-Glc-α(1→3)-Glc), linked as in its native Glc3Man9GlcNAc2 substrate. Kojibiose, the disaccharide Glc-α(1→2)-Glc, acts as a weak inhibitor on plant, animal, and yeast MOGS.
MOGS also acts to lesser extent on the corresponding glycolipids and glycopeptides.
