 | ||
Symbol UNQ6490, PRO21339, (LOC389102, YPLR6490) | ||
LOC105377021 is a protein which in humans is encoded by the LOC105377021 gene. LOC105377021 exhibits expressional pathology related to breast cancer, specifically triple negative breast cancer. LOC105377021 contains a serine rich region in addition to predicted alpha helix motifs.
Contents
- Gene and mRNA
- Protein Primary Structure
- Protein Secondary Structure
- Protein Tertiary Structure
- Protein Modifications and Localization
- Basic Expression and Breast Cancer
- Brain Tissue Expression
- Orthologs
- Pace of Evolution
- Single Nucleotide Polymorphisms
- Promoter and Gene Regulation
- References
Function: The explicit function is unknown, however based on bioinformatic data presented herein, LOC105377021 is estimated to play a maintenance role in the nucleus of the cell.
Gene and mRNA
LOC105377021 localizes to Homo sapiens chromosome three (3p2; antisense strand), approximate to the reading frame of TRIM71. The corresponding gene has 2,473 nucleotides. There is one exon in the LOC105377021p mRNA. There is no predicted alternative splicing on the NCBI gene database.
Protein Primary Structure
The figure below shows the basic primary protein structure, with N-terminus and C-terminus in their respective annotations. The orange domain is a predicted nuclear localization sequence, while the blue domain is the remainder of the LOC105377021 exon.
Protein Secondary Structure
According to Ali2D (a multiple sequence alignment structural predictor for proteins), LOC105377021 is predicted to form mostly alpha helix (see red highlight, blue highlight is for Beta Sheet).
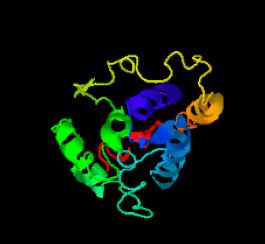
Protein Tertiary Structure
LOC105377021 has a prominent, C-terminus repeat of serine residues, potentially for disulfide bonding. One disulfide bond (139-148) was predicted by DISULFIDE software. Additionally, the I TASSER profile shows several alpha helices in a variety of different colors, in addition to potential turn motifs (see I TASSER 3D Prediction of LOC105377021).
Protein Modifications and Localization
A predicted protein modification of LOC105377021 is phosphorylation, with sites throughout the protein, including the serine rich construct near the C-terminus of the protein. In addition, there is predicted evidence of O-Linked β-N-acetylglucosamine supplements in the C terminal region. There is predicted evidence for a nuclear localization sequence oriented at the N-terminal, provided by PSORT with partial support by PHOBIUS software.
Basic Expression and Breast Cancer
Compared to the average expression of human protein, LOC105377021 is expressed at 0.9%, which is classified as low. In humans: cranial, intestinal, ovarian, renal, and testicular tissues corroborate this trend.
Microarray data posits the expression of LOC105377021 in certain breast cancer tissues, including metastases to lymphatic and lung tissue. There is potential evidence for higher expression of LOC105377021 during Triple Negative Breast Cancer, which overshadows normal secretion levels for said protein. The figure below shows a potential trend line for this pattern (shown in green, with the triple negative microarray on the left). As the figure legend states, the red bars refer to the left axis for sample counts, whereas the blue dots show the percentage of LOC105377021 expression within each sample (the right axis).
This photo is courtesy of NCBI Geo Profiles Accession GDS4069.
Brain Tissue Expression
Seven key brain tissues express LOC105377021 according to an Allen Brain Atlas probe. The temporal lobe, parietal lobe, cingulate gyrus, parahippocampal gyrus, and insula are five overarching regions of the seven brain tissues where expression was highlighted. The annotated figures below serve as fairly holistic representations of cranial expression in the context of LOC105377021. Light blue shaded regions posit more dense expression of LOC105377021, where as darker green and brighter red show less and least amounts of expression respectively. All seven expression areas, including the middle temporal gyrus, the short insular gyrus, the postcentral gyrus, the cingulate gyrus, the inferior temporal gyrus, the parahippocampal gyrus, and the superior temporal gyrus are depicted in Allen Brain Atlas profiles below.
These photos are courtesy of the Allen Brain Atlas.
Orthologs
The Basic Local Alignment Sequence Tool (BLAST) shows that LOC105377021 orthologs are largely homogenous and mammalian. Important orthologs are summarized into three categories: primates, aquatic mammals, and ferrets/ferret-like animals. Pongo abelii and Tursiops truncatus are the most distant and related orthologs respectively. The river dolphin is the first ortholog to detach from the 80% plus similarity cohort. The following includes a list of select orthologs found:
Pace of Evolution
The pace of evolution of LOC105377021 upon its inception (in humans) is modeled to be slow. This speed is relative to cytochrome c 6A1 and Alpha fibrinogen using corrected divergence methods.
The corrected divergences graph above shows three lines: Alpha Fibrinogen in Red, LOC Ortholog (aka LOC105377021) in blue, and Cytochrome c 6A1 in green. These lines associate with evolutionary pace in LOC105377021, as tested using a corrected divergence genomic analysis.
Single Nucleotide Polymorphisms
The following diagram shows single nucleotide polymorphisms (SNP's) in various regions of the protein. SNP's are highlighted green, with SNP coding on the right hand coding for switches in amino acids.
Promoter and Gene Regulation
According to Genomatix, LOC389102 (synonym to LOC105377021) is proximate to a 601 base pair promoter and a 5'UTR 129 base pairs long consecutively. Genomatix predicts several transcription factors in general. Two select factors predicted include Gli3 and E2F1.
