 | ||
isomiR is a term created by Morin et al. to refer to those sequences that have variations with respect to the reference MiRNA sequence. miRBase is considered to be the gold-standard miRNA database—it stores miRNA sequences detected by thousand of experiments. In this database each miRNA is associated with a miRNA precursor and with one or two mature miRNA (-5p and -3p). Similarly, VIRmiRNA is the only available resource for miRNA in the field of virus. It not only contains viral miRNAs but also harbors their targets. It also provides extensive list of anti-viral microRNAs encoded by human to provide them immune defense against viral infections. Currently this resource harbors 9133 records; out of which there are 1308 experimentally validated viral miRNAs from 44 viruses In the past it had always been said that the same miRNA precursor generates the same miRNA sequences. However, the advent of deep sequencing has now allowed researchers to detect a huge variability in miRNA biogenesis, meaning that from the same miRNA precursor many different sequences can be generated potentially have different targets, or even lead to opposite changes in mRNA expression. It has been found that isomiR expression profiles can also exhibit race, population, and gender dependencies.
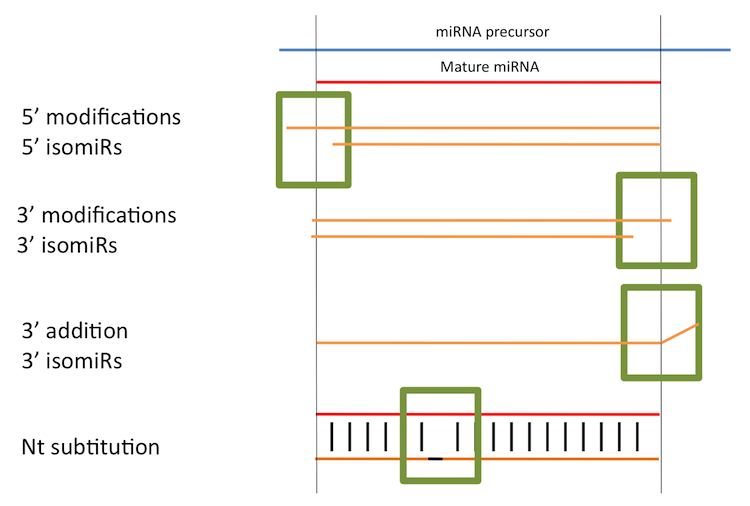
There are four main variation types:
Biogenesis
The advent of sequencing has permitted scientists to elucidate a huge landscape of new miRNAs, to increase our knowledge of the biogenesis involved and to discover putative post-transcriptional editing processes in miRNAs ignored until now. These processes mostly generate variations of the current miRNAs that are annotated in miRBase in the 3’ and 5’ terminus and in minor frequencies, nucleotide substitution along the miRNA length,. The variations are mainly generated by a shift of Drosha and Dicer in the cleavage site, but also by nucleotide additions at the 3’-end, resulting new sequences different from the annotated miRNA. These were named "isomiRs" by Morin et al., 2008. IsomiRs have been well established along different species in metazoa and deeply described for the first time in human stem cells and human brain samples. Moreover, it has been proven that isomiRs are not caused by RNA degradation during sample preparation for next generation sequencing. Some studies have tried to explain the miRNA diversity by structural bases of precursors but without clear results. The functionality of adenylation or uridynilation at the 3’end (3’addition isomiRs) has been related to alterations in the miRNA-3’-UTR stability. Furthermore, isomiRs have been detected deregulated in D. melanogaster development and differential expressed during Hippoglossus hippoglossus L. early development, suggesting a biologically relevant function.
