Possible time of origin 112,200 to 188,000 YBP | Descendants L0a'b'f'k, L0d | |
 | ||
Defining mutations 263!, 1048, 3516A, 5442, 6185, 9042, 9347, 10589, 12007, 12720 | ||
Haplogroup L0 is a human mitochondrial DNA (mtDNA) haplogroup.
Contents
Origin
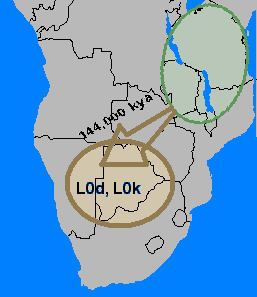
L0 is one of two branches from the most recent common ancestor (MRCA) for the shared human maternal lineage. The haplogroup consists of five main branches (L0a, L0b, L0d, L0f, L0k). Four of them were originally classified into clades L1 as L1a, L1d, L1f, and L1k.
In 2014, ancient DNA analysis of a 2,330 year old male forager's skeleton in Southern Africa found that the specimen belonged to the L0d2c1c mtDNA subclade. This maternal haplogroup is today most closely associated with the Ju, a subgroup of the indigenous San people, which points to population continuity in the region. In 2016, a Late Iron Age desiccated mummy from the Tuli region in northern Botswana was also found to belong to haplogroup L0.
Distribution
L0 is found most commonly in the Sub-Saharan Africa. It reaches its highest frequency in the Khoisan people at 73%. Some of the higher frequencies are: Namibia (!Xun) 79%, South Africa (Khwe/!Xun) 83%, and Botswana (!Kung) 100%.
Haplogroup L0d is the most divergent ("ancient") haplogroup of global mitochondrial DNA haplogroups. It is found at highest frequencies in the Khoisan groups of Southern Africa. L0d is also commonly found in the Coloured population of South Africa and frequencies range from 60% to 71%. This illustrates the massive maternal contribution of Khoisan people to the Coloured population of South Africa.
Haplogroups L0k is the second most common haplogroup in the Khoisan groups (following L0d) and is largely restricted to the Khoisan. Interestingly, while the Khoisan associated L0d haplogroup were found in high frequencies in the Coloured population of South Africa, L0k were not observed in two studies involving large groups of Coloured individuals.
Haplogroup L0f is present in relatively small frequencies in Tanzania, East Africa.
Haplogroup L0a is most prevalent in South-East African populations (25% in Mozambique). Among Guineans, it has a frequency between 1% and 5%, with the Balanta group showing increased frequency of about 11%. Haplogroup L0a has a Paleolithic time depth of about 33,000 years and likely reached Guinea between 10,000 and 4,000 years ago. It also is often seen in the Mbuti and Biaka-pygmies. L0a is found in almost 25% in Hadramawt (Yemen).
Haplogroup L0b Ethiopia.
Drug and Disease Interactions
In patients who are given the drug stavudine to treat HIV, Haplogroup L0a2 is associated with a higher likelihood of peripheral neuropathy as a side effect.
Tree
This phylogenetic tree of haplogroup L0 subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation and subsequent published research.
