 | ||
The Hac1 Xbp1 intron is a non-canonical intron, spliced from bZIP-containing genes called HAC1 in Fungi and XBP1 in Metazoa. Splicing is performed independently of the spliceosome by IRE1, a kinase with endoribonuclease activity. Exons are joined by a tRNA ligase. Recognition of the intron splice sites is mediated by a base-paired secondary structure of the mRNA that forms at the exon/intron boundaries. Splicing of the Hac1/Xbp1 intron is a key regulatory step in the unfolded protein response (UPR). The Ire-mediated unconventional splicing was first described for HAC1 in S. cerevisiae.
Contents
Consensus structure
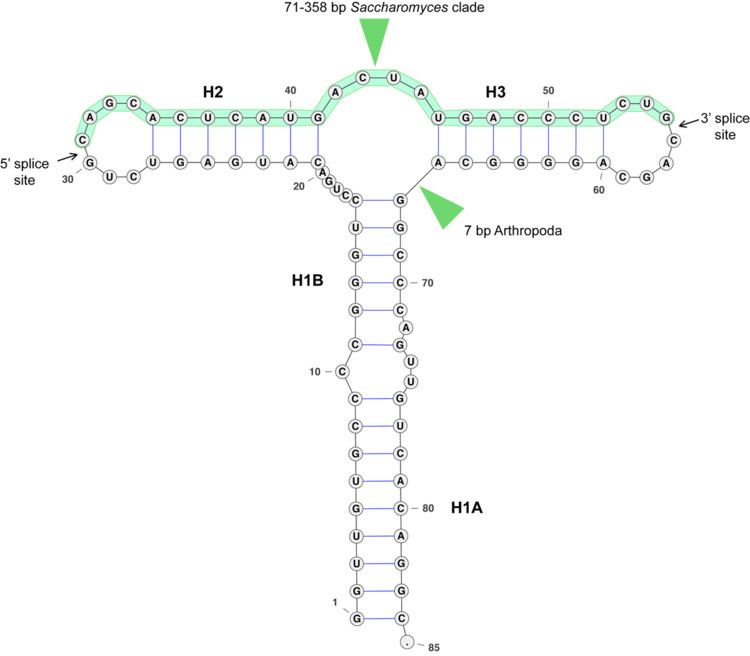
The secondary structure of the HAC1/XBP1 intron is very well conserved, and consists of two hairpins (H2 and H3) around the splice sites, and an extended hairpin (H1) that brings the splice sites together (see figure). The sequence of the intron is well conserved only around the splice sites. Non-canonical splicing motifs CNG'CNG in the loop region of H2 and H3 hairpins are conserved.
The consensus intron is very short (20, 23 or 26 nt). However, 14 yeast species have a long (>100 nt) intron in HAC1. In Saccharomyces cerevisiae the long intron pairs with the 5' UTR and stalls the ribosomes on the mRNA.
Mechanism of splicing
Environmental stress can cause proteins to misfold and aggregate. To protect from these undesirable processes, a cell can activate the unfolded protein response (UPR) pathway. Splicing of HAC1 and XBP1 transcripts is one of the highly regulated ways of activating the UPR in response to presence of unfolded proteins in the endoplasmic reticulum (ER). ER stress activates the endoribonucleolytic activity of IRE1 proteins. IRE1 recognizes splice-site motifs in HAC1 or XBP1 transcripts and cleaves them. Stem-loop structures around the splice sites and IRE1-specific sequence motifs are both necessary and sufficient for splicing to occur. The joining of exons is performed by tRNA ligase (TRL1 in Saccharomyces cerevisiae).
Intron conservation
IRE-mediated unconventional splicing of the HAC1/XBP1 intron has been confirmed experimentally in the following species:
Computational methods predict a HAC-like intron with its characteristic RNA structure in 128 out of 156 species studied. In Fungi a HAC-like intron can be found only in Ascomycota (in 52 out of 63 species analysed). All 45 vertebrate genomes analysed, 19 of Arthropoda, 7 of Nematoda, 2 of Annelida and 2 of Mollusca contain a characteristic HAC1-like structure in an open reading frame.
