RNA type Cis-regulatory element | Rfam RF01738 | |
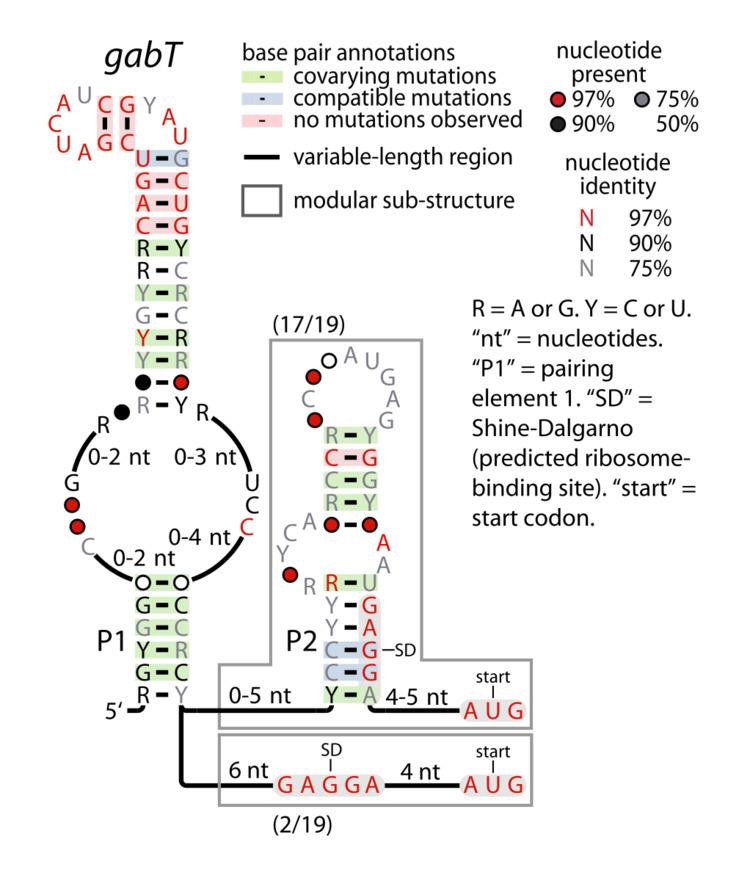 | ||
The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.
Although it is agreed that the gabT and gabD genes encode a transaminase and dehydrogenase, respectively, it is less clear what substrates they operate on. Evidence suggests that they catalyze the transamination of GABA and the dehydrogenation of the product to form succinate, an intermediate in the Krebs cycle. But it was also shown that they can produce glutarate from delta-aminovalerate (via transamination and dehydrogenation reactions) as part of the degradation of lysine. Both the gabD and gabT genes were shown, in various experiments, to be induced by metabolites such as agmatine and lysine that would fit with either of the two activities just mentioned for the products of gabD/gabT. The gabT RNA is probably not participating in this mode of regulation, unless it can regulate the upstream gene.
The predicted Shine-Dalgarno sequence (ribosome-binding site) is contained within the stem-loop called "P2" in some gabT RNAs, and nearby to the "P1" stem-loop in those gabT RNAs that do not use a P2 stem-loop. This proximity hints at a mechanism for gene regulation that gabT RNAs might effect.
