 | ||
Epitranscriptomics describes an aspect of molecular genetics, or a study thereof, that depends on biochemical modifications of RNA. By analogy to the term epigenetics, described as "functionally relevant changes to the genome that do not involve a change in the nucleotide sequence", epitranscriptomics can be defined as a functionally relevant changes to the transcriptome that do not involve a change in the ribonucleotide sequence. The epitranscriptome, therefore, is defined as the ensemble of such functionally relevant changes.
Contents
- Different Types of RNA modifications
- rRNA modifications
- tRNA modifications
- mRNA modifications
- m6A methylation
- 5 Methylctosine
- 2 O methylation
- N6 Methyladenosine m6A
- N6 Methyladenosine m6A on Alternative Splicing
- mRNA capping by NAD NADH dpCoA
- Pseudouridylation and pseudo seq identification
- References
There are several types of RNA modifications that impact gene expression. These modifications happen to all types of cellular RNA including, but not limited to, ribosomal RNA(rRNA), transfer RNA (tRNA), messenger RNA (mRNA), and small nuclear RNA (snRNA). There are more than one hundred documented RNA modifications. A database maintained by the University of Albany details each modification. The most common and well-understood mRNA modification at present is the N6-Methyladenosine (m6A), which has been observed to occur an average of three times in every mRNA molecule.
The relative youth of this field means there is still much progress to be made in characterizing all modifications to the transcriptome and elucidating their mechanisms of action. Once these questions are answered and biologists have a better sense of the amount of variation in RNA modification, the focus will turn to each modification’s biological function. This has already been investigated in a select few proteins such as adenosine deaminase, which acts on RNA (ADAR). ADAR has been shown to affect antibody production and the innate immune system as well as transcripts encoding important receptors for the central nervous system. This plurality in function has caused some scientists to speculate that the epitransciptome may be even more expansive than the better defined epigenome.
Different Types of RNA modifications
It has been recently discovered that some modification like RNA methylation, may play an active role in regulating some biological process. Also that reversible modifications might occur on tRNA, rRNA, and mRNAs to regulate gene expression and effect biological process.
Modifying an RNA molecule requires a significant amount of energy from the cell This high energy cost must mean that the cell deems these modifications to be very important. There are three known types of RNA modifications. rRNA modifications, changes that help with molecular recognition, and mRNA modifications.
rRNA modifications
rRNA changes take place in areas of high translational activity, such as the PTC(peptidyl transferase center). Some modifications include pseudouridines, 2′-O-methylations on backbone sugars, and methylated bases. It is not well known what the biological effects of these modifications are on the rRNA molecule but the hypothesis is that they help stabilize the structure and function of the ribosome especially during ribosome replication. Some functions of these modifications is that the 2' O methyl does not allow the hydrolysis of the phosphate backbone of the rRNA. It also allows for more base stacking forces. Which creates a more stable secondary structure. These modifications are required around the PTC to help with translational efficiency.
tRNA modifications
The known purposes for this kind of modification is to allow for a bigger difference between the tRNA molecules so that tRNAMet is distinguished from elongator tRNAMet and tRNA stability. Some studies have shown the modifications of tRNA can be a dynamic and adaptive to the changes for the environment. Examples include the tRNA methyltranserfase (Trm4) which methylates a cysteine group in response to the depletion of nutrients in the body.
mRNA modifications
Because mRNA is the essential code that changes our biological information from DNA to protein, through transcription and translation, the epitranscriptomic modifications that occur on the mRNA must be nonmutagenic and thus most modifications are methylations. The four sites on mRNA for methylation are N7-methylguanine (at the 5′ cap), N6-methyl adenosine, 5-methylcytosine, and 2′-O-methylation. The cap in mRNA is very important it helps the to recognition by cap-binding proteins to initiate translation, and prevents the degradation of mRNA in the cytosol.
m6A methylation
The most common modification of eukaryotic mRNA. m6A is applied by a m6A methyltransferase complex post-transcriptionally . It was only until recent years that scientists have been able to examine and try to identify the purpose of the mRNA modifications. It has been shown that three members of the human YTH domain family proteins show higher binding affinities to methylated mRNA. The YTH protein, YTHDF2, affects mRNA by directing methylated mRNA from the translational pool to mRNA decay sites. It was also found that the mRNA that was methylated has a shorter half-life and that it is filled with regulatory sequences. This suggests that the methylation has a big control on the affect of gene expression. Some m6A demethylases, such as FTO and AlkBH5, were discovered. Their function and knockout study reveals the affects they have on the body. FTO has correlations with body weight and disease, while Alkbh5 knockout mice have impaired fertility. These demethylations serve as proof that the m6A methylation that occurs on mRNA is very important for bodily function.
5-Methylctosine
An epigenetic modification that is known to occur in rRNA and tRNA, but it has been recently discovered that there are many m5C sites in mRNA. Genes that deal with energy and lipid metabolism are found near these sites which may indicate m5C involvement in this type of regulation.
2′-O-methylation
Is involved in differentiating between self and non-self mRNA. Without 2′-O-methylation activity the immune system triggers higher levels of type 1 interferon activity. It is also seen that the 2′-O-methylation occurs in plants and animals to help protect the 3' end of miRNA(microRNA) from polyadenylation.
N6-Methyladenosine (m6A )
m6A describes the methylation of the nitrogen at position 6 of the adenosine base within mRNA. Discovered in 1974, m6A is the most abundant eukaryotic mRNA modification. The term "epitranscriptome" was coined following transcriptome-wide mappings of m6A sites, but does not necessarily exclude other post-transcriptional mRNA modifications.
m6A methylation regulates nuclear export of mature mRNA and mRNA stability. How, and in response to what stimulus, the cell endogeneously regulates the level of m6A methylation remains unclear at present.
This m6A modification has a notable effect on translational dynamics. As expected, because it is just a modified adenosine base, m6A base-pairs with uridine during decoding. However, the adenosine’s methylation hinders tRNA accommodation and translation elongation. When an m6A-modified codon interacts with its cognate tRNA, it acts more like a near-cognate codon interaction instead of the cognate codon interaction. This can be seen in that the delay in the tRNA accommodation, which is dependent upon both the position of the m6A in the mRNA codons and on how accurate the translation is. To summarize, translation-elongation dynamics are slower for codons with m6A and different locations of these modified nucleotides in the mRNA codons affect decoding dynamics in different ways.
The m6A modification affects tRNA incorporation’s distinct steps in different ways. It slows down GTP hydrolysis by EF-Tu 12-fold, delays tRNA accommodation two-fold, and it slows down peptidyl transfer two-fold. It also causes a 1.5-fold increase in the amount of GTP hydrolyzed per peptidyl transfer, which indicates that a lot of proofreading is required. Overall, this m6A modification leads to a kinetic loss of a factor of 18.
N6-Methyladenosine (m6A) on Alternative Splicing
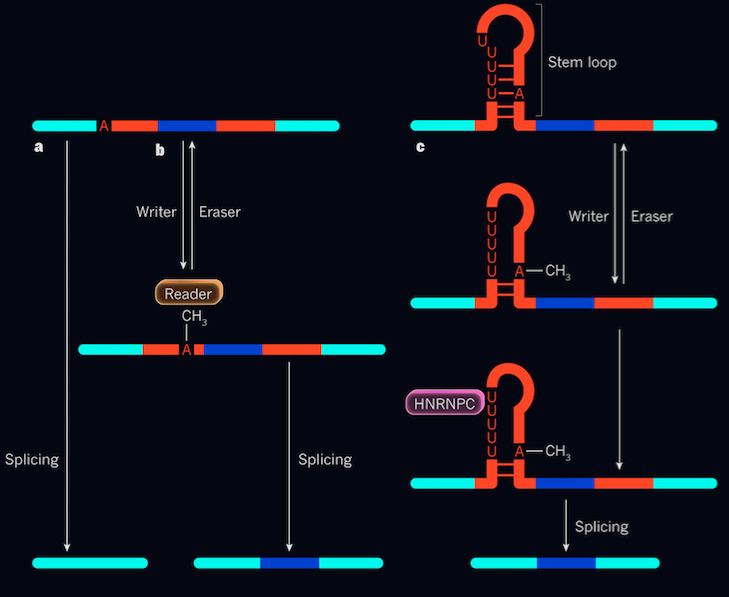
The terms “eraser” and “reader” have been associated with RNA modification. “Eraser” is a general term to describe an enzyme that de-methylates m6A. Changes that mutate the gene encoding the “eraser” enzyme lead to obesity and cancer. “Reader” proteins are involved in gene expression where there are abundant m6A; “reader” proteins have a higher propensity to bind with greater affinity, while the de-methylated form has been reported to have a decreased binding affinity.
mRNA are subject to layers of regulatory gene expression. One known mechanism involves the formation of RNA stem-loops. Stem-loops occur when complementary bases within a single-stranded RNA molecule form Watson-Crick base pairs on the stem while forming an unpaired end or loop. Stem-loops do not have one definite function, but a plethora of functions. In the case of the m6A regulatory mechanism, it is involved in alternative splicing. These stem and loop structures are subject to alterations regarding changes in pH, temperature, ion concentrations, binding nature of proteins and also nucleic acids.
m6A has been observed to be located within the loops opposite of the HNRNPC binding site. HNRNPC is single-stranded RNA binding protein where it participates in post-transcriptional regulation, specifically alternative splicing. HNRNPC protein binds to its site (uridine rich region on the stem loop) when methylated adenosine is present.The HNRNPC binding site on the mRNA consists of an abundance of uridine nucleotides. Methylated adenosine residues destabilize the hairpin structure, elongating the uridine nucleotide stretch, causing the binding site to be more accessible for efficient HNRNPC protein binding.
Evidence supporting this claim identified that decreased m6A levels in the transcriptome lead to significantly reduced HNRNPC binding, thus alternative splicing is co-regulated by methylation and HNRNPC binding activity. However, the m6A modification does not directly cause protein binding. Rather, it alters the loop structure to regulate gene expression by acting as a switch that exposes the HNRNPC region. It is essentially a two-step, co-regulatory mechanism prevalent in biochemistry in controlling alternative splicing. Note that demethylase enzyme can indeed “erase” the methyl group, thus inhibiting alternative splicing which is the reverse of the two-step regulation mechanism.
mRNA capping by NAD+, NADH & dpCoA
mRNA capping in eukaryotes have shown to exist and have been studied extensively regarding the 5’ 7-methylguanylate cap and its poly-A tail. It provides RNA stability, processing, localization and translational efficiency. Recently, researchers have proven a similar mechanism in RNA pre-processing in prokaryotes. In prokaryotic (E.coli) mRNA, the 5’ is capped with nicotinamide adenine dinucleotide (NAD+ or NADH) or 3’-desphospho-coenzyme A (dpCoA). It was previously thought that the NAD+,NADH, dpCoA modification occurs after transcription analogously with the 7-methylguanylate cap; however, recently it has been shown that modifications are incorporated during transcription initiation by acting as a non-canonical initiating nucleotides (NCIN) for de novo transcription by cellular RNA polymerase. RppH and NudC pyrophosphohydrolases cleaves specific phosphate bonds to eliminate the cap modification. ATP was initially known to cap an RNA product in prokaryotes while NAD+, NADH and dpCoA was still being studied. RppH specifically cleaves 5’-triphosphate and 5’-diphosphate RNAs to 5’-monophosphate RNA products which only targets the ATP capped RNAs. On the other hand, NudC cleaves 5’-(NADH,NAD+,dpCoA) capped RNAs to 5’-monophosphate RNAs but does not cleave ATP capped RNAs. This discovery of two specific hydrolases that target specific products lead to the discovery of NCIN-mediated transcription. Both prokaryotes and eukaryotes transcripts have been observed with x-ray crystallography to have a NCIN-capped RNA product in addition to being observed under in vivo condition. The efficiency of NCIN capping is largely influenced by promoter sequence especially the -35 box and -10 box upstream of the transcription start site. Studies so far have defined the mechanism and structural basis of NCIN mediated capping but the function of these caps are still to be discovered. Recent consensus is that NCIN-mediated ab initio capping occurs in all organisms.
Pseudouridylation and pseudo-seq identification
Pseudouridine is a modified nucleoside found within non-coding RNAs (ncRNA). It increases the function of tRNA and rRNA by stabilizing the structure. Though mRNAs are not known for containing pseudouridine, the artificial process of pseudouridylation has an affect on the function of mRNA: it changes the genetic code by making non-canonical base pairing possible in the ribosome decoding center. Pseudouridine also provides more hydrogen bonding opportunity and makes the RNA backbone more rigid.
A certain paper looks at pseudouridylation in yeast and human RNAs using pseudo-seq, a process that utilizes a single-nucleotide-resolution method for pseudouridine identification. It identifies the known modification sites as well as other sites in ncRNAs in addition to the many pseudouridylated sites in mRNA. Other methods outside os pseudo-seq that have been utilized for pseudouridine identification and detection range from pseudouridine-seq, PSI-seq, and CeU-seq.
There are more than 100 classes of RNA modifications that have been found, a majority of which are in tRNA and rRNA while only three modified nucleotides have been discovered inside a coding sequence of mRNA: m6A, 5-methylcytosine (m5C), and inosine. The relevance of pseudouridine in rRNA has been documented where pseudouridylation is required for ribosome biogenesis and translational accuracy. The importance of pseudouridylation additionally seen in snRNA where specific pseudouridine residues have been located and identified as a needed component of pre-mRNA splicing. Previous research has focused specifically at the modifications of U2 spliceosomal RNA which are required for snRNP assembly and pre-mRNA splicing. U2 in comparison to other spliceosomal snRNAs has the most extensive modifications, including 13 pseudouridines as well as a 5' trim ethyl guanosine (TMG) cap and others. It is a mixture of modifications involving that of the pseudouridine residues that assist in the complete assembly of the 17S U2 snRNP particle needed for splicing.
Pseudouridylation has been found to be inducible in response to stress and differentiation in the cell, giving reason to believe that pseudouridylation may act as an important regulator mechanism for RNA function. Much of the regulation in regards to pseudouridylation is regulated through the environment. In yeast this may be nutrient deprivation and in humans it is the serum starvation. When looking at yeast, research has utilized perturbing pseudouridine synthases deletion strains grown to high density and identified mRNA targets for each PUS protein. Results came back showing that most mRNA targets showed increase modification during post-diauxic growth. The pseudo-seq method identified 96 pseudouridines in 89 mRNAs, similar to yeast the growth of pseudouridine was regulated by cellular growth state. This approach provides an analysis of RNA pseudouridylation with single-nucleotide resolution and shows endogenous mRNAs are specifically pseudouridylated in a highly regulated manner in yeast and human cells. mRNA pseudouridylation could also bring a change in translation initiation efficiency, RNA localization, and other processes all cause pseuduridine stabilizes RNA structure.
