InterPro IPR011648 PDB RCSB PDB; PDBe; PDBj | Pfam PF07688 Pfam structures PDBsum structure summary | |
 | ||
In molecular biology, the cyanobacterial clock proteins are the main circadian regulator in cyanobacteria. The cyanobacterial clock proteins comprise three proteins: KaiA, KaiB and KaiC. The kaiABC complex may act as a promoter-nonspecific transcription regulator that represses transcription, possibly by acting on the state of chromosome compaction.
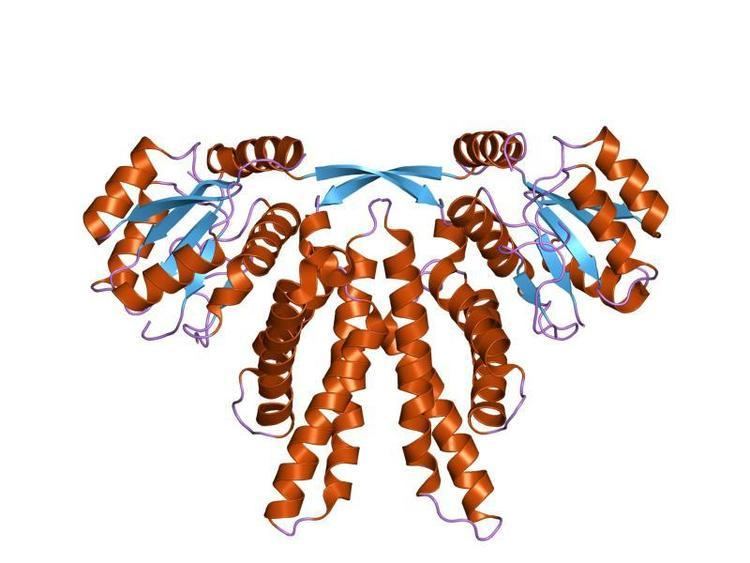
In the complex, KaiA enhances the phosphorylation status of kaiC. In contrast, the presence of kaiB in the complex decreases the phosphorylation status of kaiC, suggesting that kaiB acts by antagonising the interaction between kaiA and kaiC. The activity of KaiA activates kaiBC expression, while KaiC represses it. The overall fold of the KaiA monomer is that of a four-helix bundle, which forms a dimer in the known structure. KaiA functions as a homodimer. Each monomer is composed of three functional domains: the N-terminal amplitude-amplifier domain, the central period-adjuster domain and the C-terminal clock-oscillator domain. The N-terminal domain of KaiA, from cyanobacteria, acts as a pseudo-receiver domain, but lacks the conserved aspartyl residue required for phosphotransfer in response regulators. The C-terminal domain is responsible for dimer formation, binding to KaiC, enhancing KaiC phosphorylation and generating the circadian oscillations. The KaiA protein from Anabaena sp. (strain PCC 7120) lacks the N-terminal CheY-like domain.
KaiB adopts an alpha-beta meander motif and is found to be a dimer or a tetramer.
KaiC belongs to a larger family of proteins; it performs autophosphorylation and acts as its own transcriptional repressor. It binds ATP.
Also in the KaiC family is RadA/Sms, a highly conserved eubacterial protein that shares sequence similarity with both RecA strand transferase and lon protease. The RadA/Sms family are probable ATP-dependent proteases involved in both DNA repair and degradation of proteins, peptides, glycopeptides. They are classified in as non-peptidase homologues and unassigned peptidases in MEROPS peptidase family S16 (lon protease family, clan SJ). RadA/Sms is involved in recombination and recombinational repair, most likely involving the stabilisation or processing of branched DNA molecules or blocked replication forks because of its genetic redundancy with RecG and RuvABC.
