 | ||
Cell lists (also sometimes referred to as cell linked-lists) are a tool for finding all atom pairs within a given cut-off distance of each other in molecular dynamics simulations. These pairs are needed to compute the short-range non-bonded interactions in a system, such as Van der Waals forces or the short-range part of the electrostatic interaction when using Ewald summation.
Contents
Algorithm
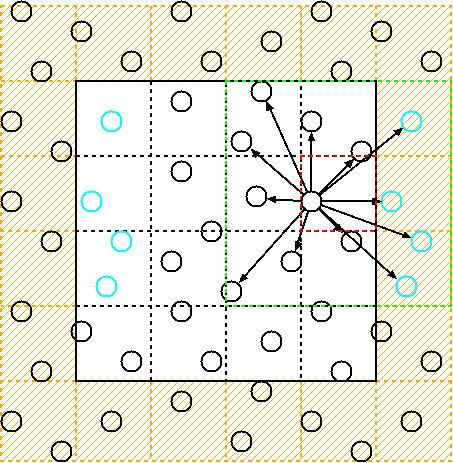
Cell lists work by subdividing the simulation domain into cells with an edge length greater than or equal to the cut-off radius of the interaction to be computed. The particles are sorted into these cells and the interactions are computed between particles in the same or neighbouring cells.
In its most basic form, the non-bonded interactions for a cut-off distance
Since the cell length is at least
Given a simulation with
Periodic boundary conditions
In most simulations, periodic boundary conditions are used to avoid imposing artificial boundary conditions. Using cell lists, these boundaries can be implemented in two ways.
Ghost cells
In the ghost cells approach, the simulation box is wrapped in an additional layer of cells. These cells contain periodically wrapped copies of the corresponding simulation cells inside the domain.
Although the data—and usually also the computational cost—is doubled for interactions over the periodic boundary, this approach has the advantage of being straightforward to implement and very easy to parallelize, since cells will only interact with their geographical neighbours.
Periodic wrapping
Instead of creating ghost cells, cell pairs that interact over a periodic boundary can also use a periodic correction vector
This approach, although more efficient than using ghost cells, is less straightforward to implement (the cell pairs need to be identified over the periodic boundaries and the vector
Improvements
Despite reducing the computational cost of finding all pairs within a given cut-off distance from
Consider a computational cell with edge length equal to the cut-off radius
One way of overcoming this inefficiency is to partition the domain into cells of edge length smaller than
Another approach is outlined and tested in Gonnet, in which the particles are first sorted along the axis connecting the cell centers. This approach generates only about 40% spurious pairwise distance computations, yet carries an additional cost due to sorting the particles.
