Entrez 11151 | Ensembl ENSG00000102879 | |
 | ||
Aliases CORO1A, CLABP, CLIPINA, HCORO1, IMD8, TACO, p57, coronin 1A External IDs MGI: 1345961 HomoloGene: 6545 GeneCards: CORO1A | ||
Coronin-1A is a protein that in humans is encoded by the CORO1A gene. It has been implicated in both T-cell mediated immunity and mitochondrial apoptosis. In a recent genome-wide longevity study, its expression levels were found to be negatively associated both with age at the time of blood sample and the survival time after blood draw.
Contents
Discovery
The Coronin protein family was discovered in 1991 by Eugenio L. Hostos. Hostos used a cytoskeletal preparation called the “contracted propeller” that efficiently helped with the purification of cytoskeletal proteins. This technique allowed him to precipitate actomyosin components together with the desired proteins.
These protein were named Corona, which is the Latin word for crown, because of the crown-like shape that it forms when making contact with the surface of the cell. Coronin-1a has been the most researched one due to its complexity and intriguing structural components. After research, it was determined that Coronin-1a serves as actin binding facilitator when reacted with K-glutamate. The anion K + and glutamate were used because of it similarity to the environment inside the cell, allowing Coronin-1a to bind to F-actin.
Later on, the complementary DNA (cDNA) of Coronin-1a was cloned in an expression library, this led to the conclusion that Coronin-1a has very similar structure to the beta (β) subunits of the G proteins (Gβ). Therefore, it was established that Coronin-1a has five WD motif repeats, and this repeats seven times forming a propeller like structure.
In the cell, Coronin-1a serves as an auxiliary to many cytoskeletal process that involve actin. It was concluded that Coronin-1a is known to affect the “cytoskeletal reorganization” as well “actin dynamics” together with other protein.
Phylogeny
The Coronin family is composed of twelve subfamilies which include: seven subfamilies that fall under vertebrates and five subfamilies that are composed of meteozas, fungi and amoeba.
The evolutionary Coronin family subfamilies have been grouped by its similarities and relationships between the different proteins. As we can see Coronin-1a (also referenced as CORO1A, Coronin 4 and CRN4) has been found in 19 vertebrates.
Function
The Coronin family is composed of twelve subfamilies which include: seven subfamilies that fall under vertebrates and five subfamilies that are composed of meteozas, fungi and amoeba.
Coronin-1a has been found in the cell cortex of macrophages, which are white blood cells, helping with a process called phagocytosis.The model on Figure 3 shows Coronin-1a’s involvement in macrophages. When the cell is at rest,Coronin-1a is spread out throughout the cytoplasm and the cell cortex. Therefore, when a pathogen enters the cell, Coronin-1a binds to phagosomal membrane making sure of the binding and activation of calcinuerin, this resulting in a stop of fusion lysosomes with phagosomes. In other words, if Coronin-1a is removed and calcinuerin is inhibited then it allows the initiation of the fusion of phagosomes with lysosome and the killing of mycobacteria.
The phylogenetic tree of the Coronin family it is quiet broad. The same way that Coronin-1a helps with the reorganization of the cytoskeleton and dynamic activity with other proteins in vertebrates, Coronin can also be seen in non-vertebrates for example the Toxoplasma gondii Coronin (also known as TgCor).
Toxoplasma gondii Coronin (TgCor) binds to F-actin and it accelerates the actin polymerization process. It also prevents incursions and exits. As well as every other coronin, TgCor is an actin binding protein, it delocalizes to the posterior side of invading parasites and blocks them from leaving.
Structure
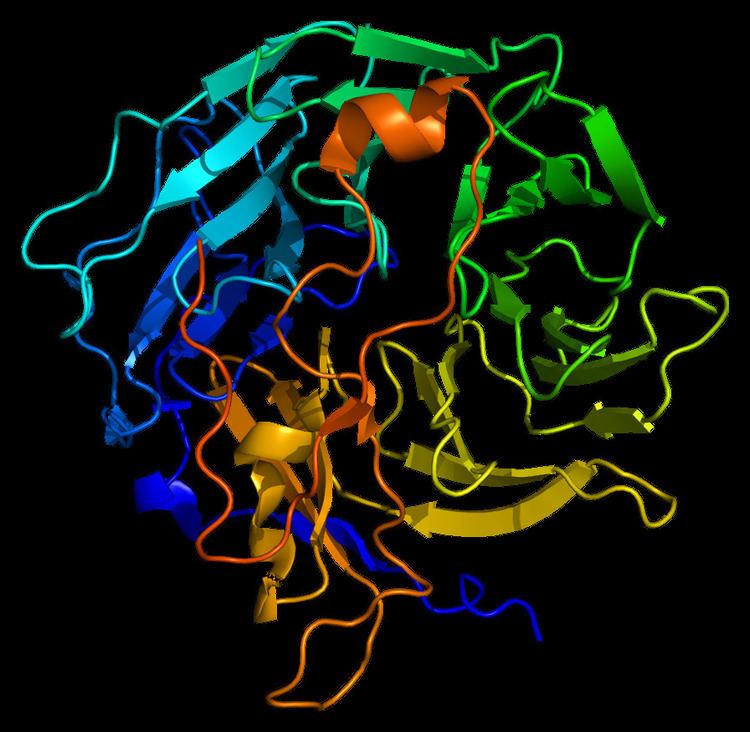
The structure of Coronin-1A is made out of five WD repeats, and this motifs repeat seven time forming a propeller like structures.
The new ribbon visualization of the secondary structure of Coronin-1a. In model A, is the front view of Coronin-1a, the secondary structure allows you to clearly see the parallel beta sheets moving towards the bottom of the structure. Model B, is the side view of the protein which shows the turns and the coils between the beta sheets. From this pictures we are able to see that the alpha helix and helix strands are concentrated at the bottom of the protein.
Coronin-1a was input into Database of Secondary Structure Program (DSSP), where the PDB database entered and a secondary structure panel is designed where one is clearly able to see the seven repeat that makes the propeller. Also, it displays the amino acid sequence of Coronin-1a. The yellow arrows mean the beta strands, the purple loops are the turns, the black lines means empty meaning that there was no secondary structure assigned, the light pink is 3/10-helix is formed, royal blue line is a bend and finally the red helix signifies the alpha helices.
