Group Group II (ssDNA) Scientific name Begomovirus Rank Genus | ||
 | ||
Lower classifications Tomato yellow leaf curl virus, Cotton leaf curl virus, Mungbean yellow mosaic vi, African cassava mosaic vi | ||
The genus Begomovirus is a genus of viruses, in the family Geminiviridae. They are plant viruses that as a group have a very wide host range, infecting dicotyledonous plants. Worldwide they are responsible for a considerable amount of economic damage to many important crops such as tomatoes, beans, squash, cassava and cotton. There are currently 322 species in this genus including the type species Bean golden yellow mosaic virus.
Contents
Taxonomy
Group: ssDNA
Morphology

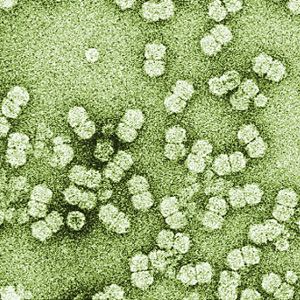
Virus particles are non-enveloped. The nucleocapsid is 38 nanometers (nm) long and 15–22 nm in diameter. While particles have basic icosahedral symmetry, they consist of two incomplete icosahedra—missing one vertex—joined together. There are 22 capsomeres per nucleocapsid.
Genome

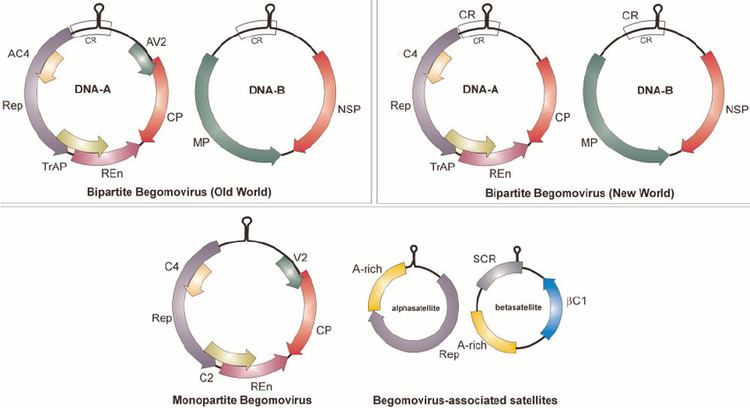
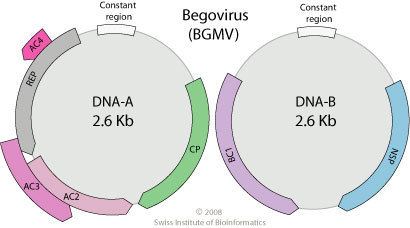
Single stranded closed circular DNA. Many begomoviruses have a bipartite genome: this means that the genome is segmented into two segments (referred to as DNA A and DNA B) that are packaged into separate particles. Both segments are generally required for successful symptomatic infection in a host cell but DNA B is dependent for its replication upon DNA A, which can in some begomoviruses apparently cause normal infections on its own.
The DNA A segment typically encodes five to six proteins including replication protein Rep, coat protein and transport and/or regulatory proteins. This component is homologous to the genomes of all other geminiviruses. The proteins endcoded on it are required for replication (Rep), control of gene expression, overcoming host defenses, encapsidation (coat protein) and insect transmission. The DNA B segment encodes two different movement proteins. These proteins have functions in intra- and intercellular movement in host plants.
The A and B components share little sequence identity with the exception of a ~200 nucleotide sequence with typically >85% identity known as the common region. This region includes an absolutely conserved (among geminiviruses) hairpin structure and repeated sequences (known as 'iterons') that are the recognition sequences for binding of the replication protein (Rep). Within this loop there is a nonanucleotide sequence (TAATATTAC) that acts as the origin (ori) of virion strand DNA replication.
Component exchange (pseudorecombination) occurs in this genus. The usual mechanism of pseudorecombination is by a process known as 'regulon grafting': the A component donates its common region by recombination to the B component being captured. This results in a new dependent interaction between two components.

The proteins in this genus may lie either on the sense strand (positive orientation) or its complement (negative orientation).
Genes

Virology
Smaller than unit length virus components—deletion mutants—are common in infections. These are known as defective interfering (di) DNAs due to their capacity to interfere with virus infection. They reduce virus DNA levels and symptom severity.
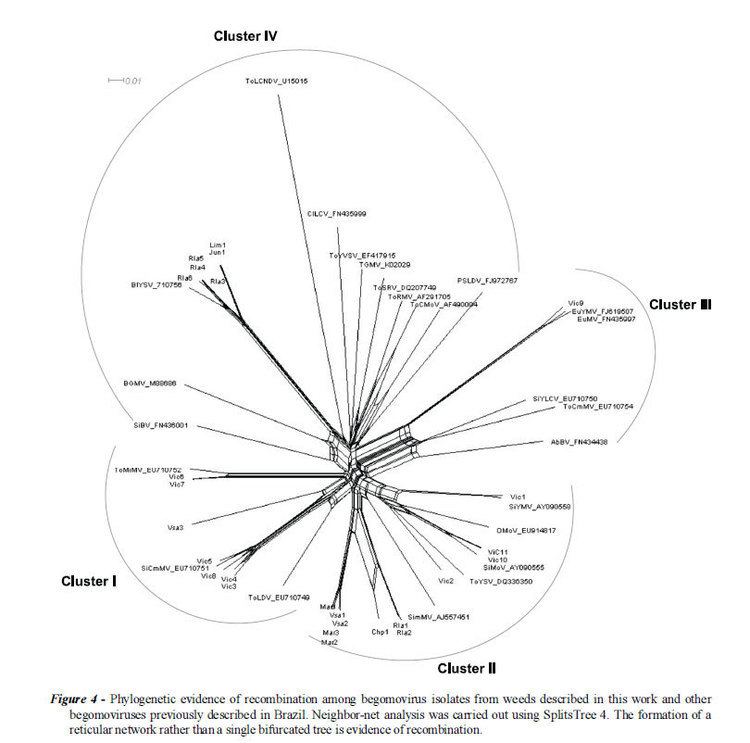
Phylogenetics
The two components of the genome have very distinct molecular evolutionary histories and likely to be under very different evolutionary pressures. The DNA B genome originated as a satellite that was captured by the monopartite progenitor of all extant bipartite begomoviruses and has subsequently evolved to become an essential genome component.
More than 133 begomovirus species having monopartite genomes are known: all originate from the Old World. No monopartite begomoviruses native to the New World have yet been identified.
Phylogenetic analysis is based on the A component. B components may be exchanged between species and may result in new species.
Analysis of the genus reveals a number of clades. The main division is between the Old and New World strains. The Old World strains can be divided into African, Indian, Japanese and other Asian clades with a small number of strains grouping outside these. The New World strains divide into Central and Southern America strains.
Along with these main groupings are a number of smaller clades. One group infecting a range of legumes originating from India and Southeast Asia (informally 'Legumovirus') and a set of viruses isolated from Ipomoea species originating from America, Asia and Europe (informally 'Sweepovirus') appear to be basal to all the other species. Two species isolated from Corchorus from Vietnam (informally 'Corchovirus') somewhat unexpectedly group with the New World species.
Transmission
The virus is obligately transmitted by an insect vector, which can be the whitefly Bemisia tabaci or can be other whiteflies. This vector allows rapid and efficient propagation of the virus because it is an indiscriminate feeder.
Diseases
Tomato yellow mosaic virus (ToYMV) is a Begomovirus, first identified in the late 1980s, that causes an infection in tomatoes. Disease is manifested in the infected plant as yellow mosaic or mottling, leaf distortion and crinkling and stunting. In Trinidad this disease in tomato is endemic and causes an estimated yield loss of 50–60%. ToYMV disease is also an economical problem in the Caribbean. Bean golden yellow mosaic virus (BGYMV) causes a serious disease in bean species within Central America, the Caribbean and southern Florida.
Additional reading
Mansoor S, Briddon RW, Zafar Y, Stanley J (March 2003). "Geminivirus disease complexes: an emerging threat". Trends Plant Sci. 8 (3): 128–34. doi:10.1016/S1360-1385(03)00007-4. PMID 12663223.
Briddon RW, Stanley J (January 2006). "Subviral agents associated with plant single-stranded DNA viruses". Virology. 344 (1): 198–210. doi:10.1016/j.virol.2005.09.042. PMID 16364750.
Sinisterra XH, McKenzie CL, Hunter WB, Powell CA, Shatters RG (May 2005). "Differential transcriptional activity of plant-pathogenic begomoviruses in their whitefly vector (Bemisia tabaci, Gennadius: Hemiptera Aleyrodidae)". J. Gen. Virol. 86 (Pt 5): 1525–32. doi:10.1099/vir.0.80665-0. PMID 15831966.
Hunter WB, Hiebert E, Webb SE, Tsai JH, Polston JE (1998). "Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera: Aleyrodidae". Plant Disease. 82 (10): 1147–51. doi:10.1094/PDIS.1998.82.10.1147.
