 | ||
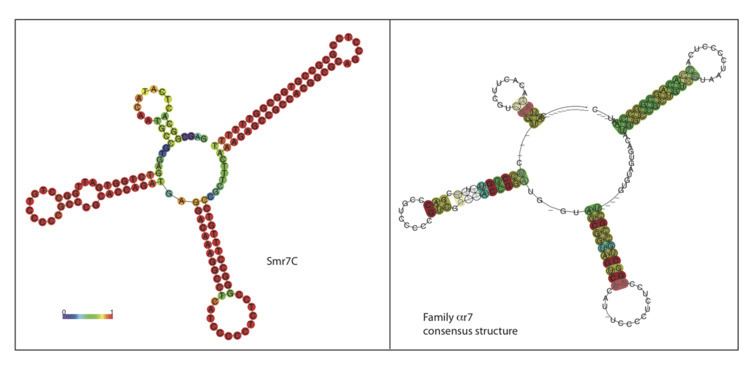
αr7 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacterial species from the order Rhizobiales. The first member of this family (Smr7C 150nt) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis identified full-length homologs in several nitrogen-fixing symbiotic rhizobia (i.e. R. leguminosarum bv.viciae, R. leguminosarum bv. trifolii, R. etli, and several Mesorhizobium species), in the plant pathogens belonging to Agrobacterium species (i.e. A. tumefaciens, A. vitis, A. radiobacter, and Agrobacterium H13) as well as in a broad spectrum of Brucella species (B. ovis, B. canis, B. abortus and B. microtis, and several biobars of B. melitensis). αr7 RNA species are 134-159 nucleotides (nt) long (Table 1) and share a well defined common secondary structure (Figure 1, Figure 2). αr7 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes (Figure 4).
Contents
Discovery and structure
Smr7C sRNA was described by del Val et al. in the intergenic regions (IGRs) of the reference S. meliloti 1021 strain (Sinorhizobium meliloti 1021). Northern hybridization experiments detected two RNA species expressed from the smr7C locus, which accumulated differentially in free-living and endosymbiotic bacteria. TAP-based 5’-RACE experiments mapped the transcription start site (TSS) to two close G residues. From the six independent sequences obtained, five mapped to residue 201,681 in the chromosome and one to the upstream residue 201,679. Furthermore, an additional 5′-RACE product could be obtained from this transcript in both TAP- and mock-treated RNA samples. The sequence of this second RACE product mapped the processing site of Smr7C to residue 201,723 nt in the S. meliloti 1021 genome. The 3’-end was assumed to be located at the 201,828 nt position matching the last residue of the consecutive stretch of Us of a bona fide Rho-independent terminator. Parallel and later studies in which Smr7C transcript is referred to as sra03 or Sm13, independently confirmed the expression this sRNA in S. melilloti and in its closely related strain 2011. Recent deep sequencing-based characterization of the small RNA fraction (50-350 nt) of S. meliloti 2021 further also confirmed the expression of Smr7C (here referred to as SmelC023), and mapped the 5’- end of the full-length transcript to the same position in the S. meliloti 1021 genome, and the 3' end to position 201,825.
The nucleotide sequence of Smr7C was initially used as query to search against the Rfam database (version 10.0;). This homology search rendered no matches to known bacterial sRNA in this database. Smr7C was next BLASTed with default parameters against all the currently available bacterial genomes (1,615 sequences at 20 April 2011;). The regions exhibiting significant homology to the query sequence (78-89% similarity) were extracted to create a Covariance Model (CM) from a seed alignment using Infernal (version1.0). This CM was used in a further search for new members of the αr7 family in the existing bacterial genomic databases (Table 1) and to create the final CM model (Figure 1).
The results were manually inspected to deduce a consensus secondary structure for the family (Figure 1 and Figure 2). The consensus structure was also independently predicted with the program locARNATE with very similar predictions. The manual inspection of the sequences found with the CM using Infernal allowed finding 26 true homolog sequences, all of them present as single chromosomal copies in the α-proteobacterial genomes. The rhizobial species encoding the closer homologs to Smr7C were: S. medicae and S. fredii, two R. leguminosarum trifolii strains (WSM2304 and WSM1325), two R. etli strains CFN 42 and CIAT 652, the reference R. leguminosarum bv. viciae 3841 strain, and the Agrobacterium species A. vitis, A. tumefaciens, A. radiobacter and A. H13 and the Mesorhizobum species loti, M. ciceri and M. BNC. All these sequences showed significant Infernal E-values (3.66E-49 - 2.93E-12) and bit-scores. The rest of the sequences found with the model showed high E-values between (1.58E-10 and 2.88E-09) but lower bit-scores and are encoded by Brucella species (B. ovis, B. canis, B. abortus, B. microtis, and several biobars of B. melitensis), and Ochrobactrum anthropi.
Expression information
Parallel studies assessed Smr7C expression in S. meliloti 1021 under different biological conditions; i.e. bacterial growth in TY, minimal medium (MM) and luteolin-MM broth and endosymbiotic bacteria (i.e. mature symbiotic alfalfa nodules). Expression of Smr7C in free-living bacteria was found to be growth-dependent, being the gene strongly down-regulated when bacteria entered the stationary phase. Interestingly, expression of SmrC7 increased ~13-fold in nodules when compared with free-living bacteria (log phase TY or MM cultures), suggesting the induction of this sRNAs during bacterial infection and/or bacteroid differentiation. SmrC7 expression has also been proved in parallel studies.
Promoter analysis
All the promoter regions of the αr7 family members examined so far are very conserved in a sequence stretch extending up to 80 bp upstream of the transcription start site of the sRNA. All closest homologous loci have recognizable σ70-dependent promoters showing a -35/-10 consensus motif CTTAGAC-n17-CTATAT, which has been previously shown to be widely conserved among several other genera in the α-subgroup of proteobacteria. To identify binding sites for other known transcription factors we used the fasta sequences provided by RegPredict, and used those position weight matrices (PSWM) provided by RegulonDB. We built PSWM for each transcription factor from the RegPredict sequences using the Consensus/Patser program, choosing the best final matrix for motif lengths between 14–30 bps a threshold average E-value < 10E-10 for each matrix was established, (see "Thresholded consensus" in gps-tools2.its.yale.edu). Moreover, we searched for conserved unknown motifs using MEME and used relaxed regular expressions (i.e. pattern matching) over all promoters of the Smr7C homologs promoters.
This studies revealed a 20 bp conserved motif in ll promoter regions, marked in orange as MEME conserved motif, in (Figure 4), but no significant similarity to known transcription factor binding sites matrices could be established.
Genomic context
All identified members of the αr7 family are trans-encoded sRNAs transcribed from independent promoters in chromosomal IGRs. Most of the neighboring genes of the seed alignment’s members were not annotated and thus were further manually curated.
The αr7 sRNAs' genomic regions of the Sinorhizobium, Rhizobium and Agrobacterium group members exhibited a great degree of conservation with the upstream and downstream genes coding for a DNA polymerase, and a MarR family transcriptional regulator, respectively. Partial synteny of the αr7 genomic regions was observed in the Mesorhizobium and Brucella group, where instead of the MarR family transcriptional regulator gene, a peptidase encoding gene was found.
Data download
Covariance Model in stockholm format can be downloaded at gps-tools2.its.yale.edu.
