 | ||
αr15 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first members of this family (smr15C1 and smrC15C2) were found tandemly arranged in the same intergenic region (IGR) of the Sinorhizobium meliloti 1021 chromosome (C). Further homology and structure conservation analysis have identified full-length Smr15C1 and Smr15C2 homologs in several nitrogen-fixing symbiotic rhizobia (i.e. R. leguminosarum bv. viciae, R. leguminosarum bv. trifolii, R. etli, and several Mesorhizobium species), in the plant pathogens belonging to Agrobacterium species (i.e. A. tumefaciens, A. vitis, A. radiobacter, and Agrobacterium H13) as well as in a broad spectrum of Brucella species (B. ovis, B. canis, B. abortus and B. microtis, and several biovars of B. melitensis). The Smr15C1 (115 nt) and Smr15C2 (121 nt) homologs are also encoded in tandem within the same IGR region of Rhizobium and Agrobacterium species, whereas in Brucella species the αr15C loci are spread in the IGRs of Chromosome I. Moreover, this analysis also identified a third αr15 loci in extrachromosomal replicons of the mentioned nitrogen-fixing α-proteobacteria and in the Chromosome II of Brucella species. αr15 RNA species are 99-121 nt long (Table 1) and share a well defined common secondary structure consisting of three stem loops (Figure 1). The transcripts of the αr15 family can be catalogued as trans-acting sRNAs encoded by independent transcription units with recognizable promoter and transcription termination signatures within intergenic regions (IGRs) of the α-proteobacterial genomes (Figure 5).
Contents
Discovery and structure
Smr15C1 y Smr15C2 sRNAs were described by del Val et al., as a result of a computational comparative genomic approach in the intergenic regions (IGRs) of the reference S. meliloti 1021 strain (http://iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime.cgi). Although the primary nucleotide sequence of Smr15C1 y Smr15C2 showed high similarity (84% identity), specific probes for each sRNA could be designed which detected transcripts of different size and expression profiles.
TAP-based 5’-RACE experiments mapped the Smr15C1 and Smr15C2 transcription start sites (TSS) in the S. meliloti 1021 genome (http://iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime.cgi). The Smr15C1 TSS was mapped to the chromosomal position 1698731 nt and the TSS of Smr15C2 to the nt 1698937. The 3’-ends were assumed to be located at the 1698617 nt and 1698817 nt respectively, matching the last residue of the consecutive Us stretch of a bona fide Rho-independent terminator (Figure 5). Parallel and later studies, in which Smr15C1 and Smr15C2 transcripts are referred to as two copies of sra41 or Sm3/Sm3', independently confirmed the expression of these sRNAs in S. melilloti and in its closely related strain 2011. Recent deep sequencing-based characterization of the small RNA fraction (50-350 nt) of S. meliloti 2011 also revealed the expression of Smr15C1 and Smr15C2, here referred to as SmelC411 and SmelC412 respectively, mapping the 5’- and 3´-ends of the full-length transcripts to essentially the same positions as del Val et al. in the S. meliloti 1021 chromosome. However, this study identified an additional TSS for Smr15C2 at position 1698948.
The nucleotide sequences of Smr15C1 and Smr15C2 were initially used as query to search against the Rfam database (version 10.0; http://www.sanger.ac.uk/Software/Rfam). This search revealed partial homology of both transcripts, restricted to the second hairpin and the Rho-independent terminator, to the RF00519 family of RNAs known as suhB (http://rfam.sanger.ac.uk/family/RF00519). However, no structural homologs of the full-length sRNAs were found in this database.
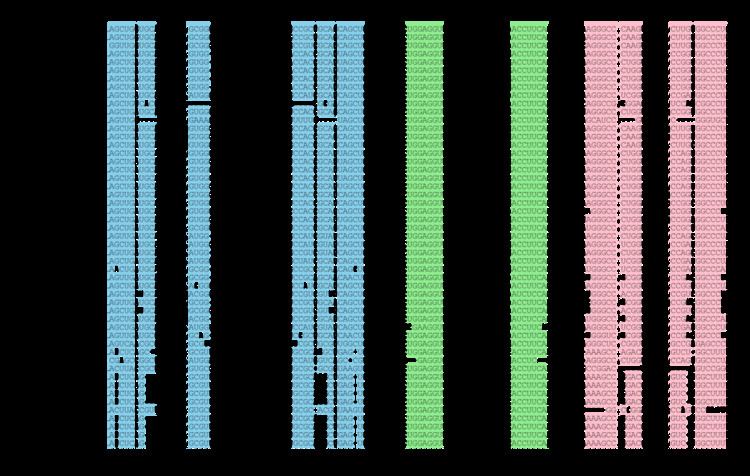
Both S.melilloti αr15 sRNAs were also BLASTed with default parameters against all the currently available bacterial genomes (1,615 sequences at 20 April 2011; http://www.ncbi.nlm.nih.gov). The regions exhibiting significant homology to the query sequence (78-89% similarity) were extracted to create a Covariance Model (CM) from a seed alignment using Infernal (version1.0) (Figure 2). This CM was used in a further search for new members of the αr15 family in the existing bacterial genomic databases.
The results were manually inspected to deduce a consensus secondary structure for the family (Figure 1 and Figure 2). The consensus structure was also independently predicted with the program locARNATE comparing the obtained predictions. The manual inspection of the sequences found with the CM using Infernal allowed finding 38 true homologues in phylogenetically related α-proteobacterial genomes. The 26 closest αr15 family members were found as tandem in the same chromosomal IGRs for the following species besides S. melilloti:
All these sequences showed significant bit scores and Infernal E-values (1.71e-28 - 2.03e-20). However, the plasmidic copies of all mentioned α-proteobacterial genomes and those αr15 members encoded by Brucella species (B. ovis, B. canis, B. abortus, B. microtis, and several biovars of B. melitensis), Ochrobactrum anthropi and Mesorhizobium lotishowed high E-values between (1e-19 and 8e-03) but very low bit-scores.
Expression and functional information
Several studies have assessed Smr15C1 and Smr15C2 expression in S. meliloti 1021 under different biological conditions; i.e. bacterial growth in TY, minimal medium (MM) and luteolin-MM broth and endosymbiotic bacteria (i.e. mature symbiotic alfalfa nodules), high salt stress, oxidative stress and cold and hot shock stresses. The results showed different expression profiles for both sRNAs, which is consistent with their organization in independent and differentially regulated transcription units within the same IGR (Figure 4 and Figure 5).
The expression of Smr15C1 and Smr15C2 in free-living bacteria was found to be growth-dependent but in an opposite manner. While Smr15C1 is accumulated in the stationary phase Smr15C2 is The expression of Smr15C1 and Smr15C2 in free-living bacteria was found to be growth-dependent but in an opposite manner. While Smr15C1 is accumulated in the stationary phase, Smr15C2 is preferentially expressed in log bacterial cultures. Additionally, Schlüter et al. recently described the up-regulation of Smr15C2 under cold shock stress, while no effects of a temperature downshift were observed in the expression of Smr15C1. Interestingly, the growth-dependent opposite expression profiles of Smr15C1 and Smr15C2, have not been observed in their Agrobacterium tumefaciens counterparts referred to as AbcR1 and AbcR2, respectively, by Wilms et al. (Atr15C1 and Atr15C2 in this work). AbcR1 and AbcR2 are induced simultaneously and both accumulate in stationary phase. This behavior agrees with the fact that AbcR1 and AbcR2 have identical promoter-like sequences, being these very similar to the one of Smr15C2, but not to the promoter sequence of Smr15C1 (see Promoter Analysis). Furthermore, a first approach to the function of the AbcR genes revealed that these sRNAs silence the GABA uptake system through the down-regulation of the corresponding ABC transporter genes in an Hfq-dependent manner. GABA is one of the plants signals recognized by rhizobacteria in some plant-bacteria interactions. Thus, these results, point to the shutting off synthesis of the GABA uptake system as a way used by A. tumefaciens to subvert the plant defense mechanism.
Recent co-inmuno precipitation experiment showed that both, Smr15C1 and Smr15C2, do bind the S. meliloti RNA chaperone Hfq, supporting also a role for these transcripts in this bacterium as trans-acting antisense riboregulators.
Promoter analysis
All αr15 loci have recognizable σ70-dependent promoters showing a -35/-10 consensus motif CTTGAC-n17-CTATAT, which has been previously shown to be widely conserved among several other genera in the α-subgroup of proteobacteria. A multiple sequence alignment of these promoter regions revealed a conserved sequence stretch extending up to 80 bp upstream of the transcription start site in all the αr15 loci with the only exceptions of the S. meliloti, S.fredii and S. medicae αr15C1 promoters.
To identify binding sites for other known transcription factors we used the fasta sequences provided by RegPredict(http://regpredict.lbl.gov/regpredict/help.html), and used those position weight matrices (PSWM) provided by RegulonDB (http://regulondb.ccg.unam.mx). We built PSWM for each transcription factor from the RegPredict sequences using the Consensus/Patser program, choosing the best final matrix for motif lengths between 14–30 bps a threshold average E-value < 10E-10 for each matrix was established, (see "Thresholded consensus" in http://gps-tools2.its.yale.edu). Moreover, we searched for conserved unknown motifs using MEME (http://meme.sdsc.edu/meme4_6_1/intro.html) and used relaxed regular expressions (i.e. pattern matching) over all αr15 homologs promoters.
These studies revealed a difference in regulation between S. melilloti, S.fredii and S. medicae αr15C1 sRNAs and the rest of the αr15 family members independently of their group of origin (Rhizobium or Agrobacterium ) and genomic location (αr15C1, αr15C2, αr15plasmid) (Figure 4).
The rest of the family members presented a very well conserved 30 bp long region between positions -36 and -75. This conserved region, was used to query databases of known transcription factor motifs with TomTom, the best matching motif was SMb20667_Rhizobiales, motif belonging to RegTransBase (http://regtransbase.lbl.gov/cgi-bin/regtransbase?page=main). The Smb20667 correspond to the binding site for a dicarboxylate metabolism regulator in Rhizobiales that belongs to the LacI family. This motif was identified clustering genes of tartrate dehydrogenase, succinate semialdehyde dehydrogenase, 3-hydroxyisobutyrate dehydrogenase and hydroxypyruvate isomerase in S. meliloti, and several Rhizobiums and it is marked in the promoter alignment figure (Figure 5) with an orange box. Moreover, another conserved sequence was found using MEME in the promoter region of the Sinorhizobium species αr15C1. This conserved region was used to query databases of known transcription factor motifs with TomTom, and the best matching motif was SMb20537_Rhizobiales (Figure 5 in red), that identifies the binding site for a sugar utilization regulator in Rhizobiales from the LacI family as well.
Genomic context
Most of the αr15 family members are trans-encoded sRNAs transcribed from independent promoters in chromosomal IGRs. Many of the αr15 members neighboring genes were not annotated, and thus they were further manually curated. As a result, we could classify the members of the family in four subgroups according to their genomic context. In the first group are the tandem αr15C1 and αr15C2 loci of the Rhizobium, Sinorhizobium and Agrobacterium species. They exhibited a great degree of conservation in the up and downstream genes, which have been predicted to code for a LysR transcriptional regulator and an AsR transcriptional regulator protein respectively (Figure 5). The only exception in this group was found for S. fredii that presented a very different genomic context. The second group includes the αr15CI1 loci in the Brucella species (additional file 2), which presented a very well conserved genomic context (Aspartate amino transferase and LysR/unknown transcriptional regulator) with partial synteny to the first group. A very different genomic context, not even partially conserved in most cases, was present in all plasmid-borne αr15 loci (additional file 1), which integrates the defined group three, where the flanking genes corresponded to ABC transporter proteins, excisonase or transposase among others. The αr15CI2 loci in the Brucella species (additional file 3) conform the group four and presented an up and downstream conserved genomic context, coding all regions for UDP-3-O-hydroxymyristoyl N-acetylglucosamine deacetylase and GTPAse cell division protein FtsZ. To the last group correspond the αr15CII loci in the Brucella group (additional file 4) where only one of the genes could be annotated always as a glycine deshidrogenase, the other sRNA flanking position was mostly coped by a hypothethical protein conserved in the Brucella group to which no domain, motif or GO functional annotation could be assigned.
Additional Files:
