 | ||
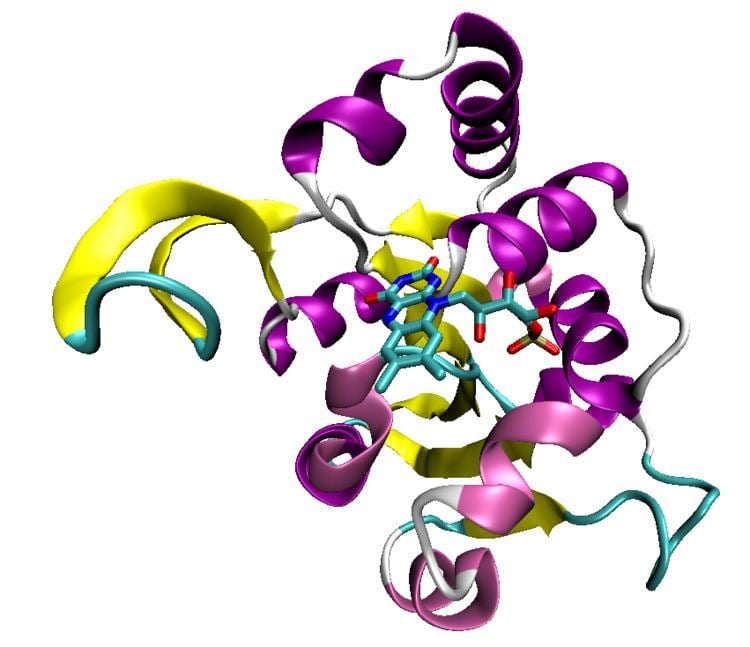
The Rossmann fold, or rather the Rao-Rossmann fold, is a protein structural motif found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. The structure is composed of up to seven mostly parallel beta strands. The first two strands are connected by an α- helix. The structures of segments between additional strands vary greatly and may include structures such as short helical segments and coils.
The motif is named for Michael Rossmann, who first pointed out that this is a frequently occurring motif in nucleotide binding proteins, such as dehydrogenases.
In 1989, Israel Hanukoglu from the Weizmann Institute of Science discovered that the consensus sequence for NADP+ binding site in some enzymes that utilize NADP+ differs from the NAD+ binding motif. This discovery was used to re-engineer coenzyme specificities of enzymes.
