 | ||
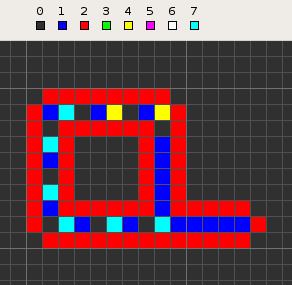
Langton's loops are a particular "species" of artificial life in a cellular automaton created in 1984 by Christopher Langton. They consist of a loop of cells containing genetic information, which flows continuously around the loop and out along an "arm" (or pseudopod), which will become the daughter loop. The "genes" instruct it to make three left turns, completing the loop, which then disconnects from its parent.
Contents
History
In 1952 John von Neumann created the first cellular automaton (CA) with the goal of creating a self-replicating machine. This automaton was necessarily very complex due to its computation- and construction-universality. In 1968 Edgar F. Codd reduced the number of states from 29 in von Neumann's CA to 8 in his. When Christopher Langton did away with the universality condition, he was able to significantly reduce the automaton's complexity. Its self-replicating loops are based on one of the simplest elements in Codd's automaton, the periodic emitter.
Specification
Langton's Loops run in a CA that has 8 states, and uses the von Neumann neighborhood with rotational symmetry. The transition table can be found here: [1].
As with Codd's CA, Langton's Loops consist of sheathed wires. The signals travel passively along the wires until they reach the open ends, when the command they carry is executed.
Colonies
Because of a particular property of the loops' "pseudopodia", they are unable to reproduce into the space occupied by another loop. Thus, once a loop is surrounded, it is incapable of reproducing, resulting in a coral-like colony with a thin layer of reproducing organisms surrounding a core of inactive "dead" organisms. Unless provided unbounded space, the colony's size will be limited. The maximum population will be asymptotic to
Encoding of the genome
The loops' genetic code is stored as a series of nonzero-zero state pairs. The standard loop's genome is illustrated in the picture at the top, and may be stated as a series of numbered states starting from the T-junction and running clockwise: 70-70-70-70-70-70-40-40. The '70' command advances the end of the wire by one cell, while the '40-40' sequence causes the left turn. State 3 is used as a temporary marker for several stages.
While the roles of states 0,1,2,3,4 and 7 are similar to Codd's CA, the remaining states 5 and 6 are used instead to mediate the loop replication process. After the loop has completed, state 5 travels counter-clockwise along the sheath of the parent loop to the next corner, causing the next arm to be produced in a different direction. State 6 temporarily joins the genome of the daughter loop and initialises the growing arm at the next corner it reaches.
The genome is used a total of six times: once to extend the pseudopod to the desired location, four times to complete the loop, and again to transfer the genome into the daughter loop. Clearly, this is dependent on the fourfold rotational symmetry of the loop; without it, the loop would be incapable of containing the information required to describe it. The same use of symmetry for genome compression is used in many biological viruses, such as the icosahedral adenovirus.
