 | ||
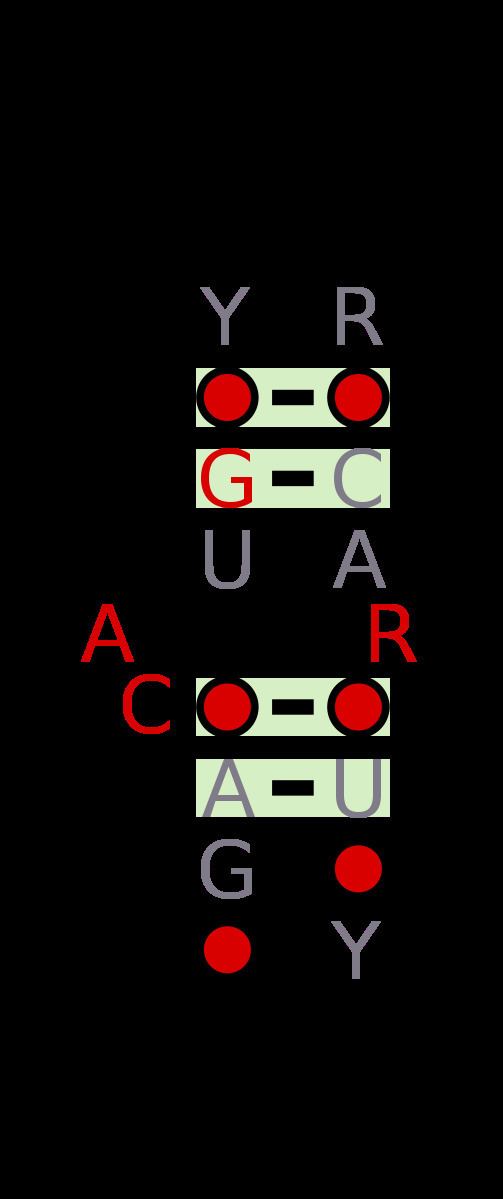
Internal-loops (also termed interior loops) in RNA are found where the double stranded RNA separates due to no Watson-Crick base pairing between the nucleotides. Internal-loops differ from Stem-loops as they occur in middle of a stretch of double stranded RNA. The non-canonicoal residues result in the double helix becoming distorted due to unwinding, unstacking and kinking.
Internal-loops can be classified as either symmetrical or asymmetrical, with some asymmetrical internal-loops, also known as bulges. Many important structural motifs are composed of internal loops such as the C-loop, the docking-elbow, kink-turns (k-turn), the right-angle, the sarcin/ricin loops (also called bulged-G motifs), the twist-up motif and the UAA/GAN internal loop motif.
