Symbol DsrA SO 0000378 | Rfam RF00014 | |
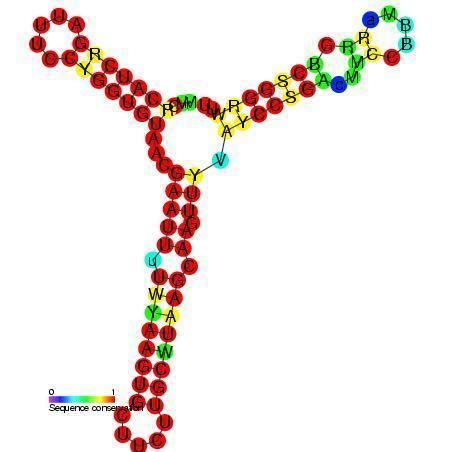 | ||
DsrA RNA is a non-coding RNA that regulates both transcription, by overcoming transcriptional silencing by the nucleoid-associated H-NS protein, and translation, by promoting efficient translation of the stress sigma factor, RpoS. These two activities of DsrA can be separated by mutation: the first of three stem-loops of the 85 nucleotide RNA is necessary for RpoS translation but not for anti-H-NS action, while the second stem-loop is essential for antisilencing and less critical for RpoS translation. The third stem-loop, which behaves as a transcription terminator, can be substituted by the trp transcription terminator without loss of either DsrA function. The sequence of the first stem-loop of DsrA is complementary with the upstream leader portion of RpoS messenger RNA, suggesting that pairing of DsrA with the RpoS message might be important for translational regulation.
There is evidence that DsrA RNA can self-assemble into nanostructures through antisense interactions of three self-complementary regions.
Targets of DsrA
There is experimental evidence to suggest that DsrA interacts with the protein-coding genes hns, rbsD, argR, ilvI and rpoS via an anti-sense mechanism.
DsrA folds into a structure with three hairpins. The second of these (nucleotides 23-60) binds to Hfq.
