 | ||
In evolutionary developmental biology, the concept of deep homology is used to describe cases where growth and differentiation processes are governed by genetic mechanisms that are homologous and deeply conserved across a wide range of species. Whereas ordinary homology is seen in the pattern of structures such as limb bones of mammals that are evidently related, deep homology can apply to groups of animals that have quite dissimilar anatomy.
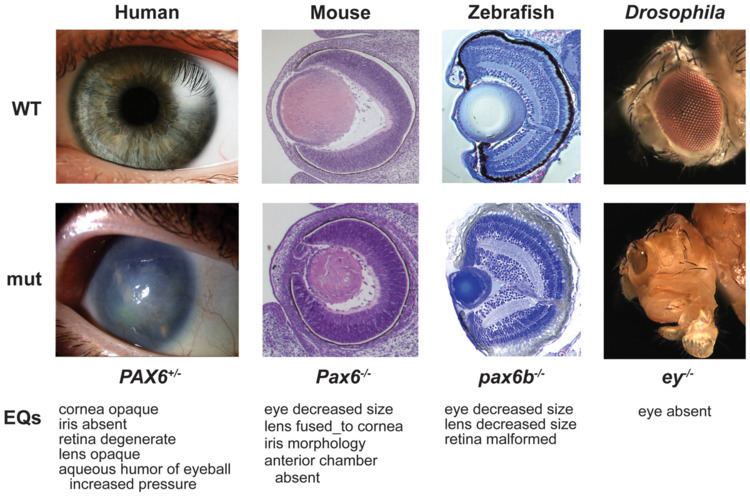
Textbook examples common to metazoa include the homeotic genes that control differentiation along major body axes, and pax genes (especially PAX6) involved in the development of the eye and other sensory organs. The deep homology applies across widely separated groups, such as in the eyes of mammals and the structurally quite different compound eyes of insects. Similarly, hox genes help to form an animal's segmentation pattern. HoxA and HoxD, that regulate finger and toe formation in mice, control the development of ray fins in zebrafish; these structures had until then been considered non-homologous. In early 2010, a team at The University of Texas at Austin led by Edward Marcotte developed an algorithm that identifies deeply homologous genetic modules in unicellular organisms, plants, and non-human animals based on phenotypes (such as traits and developmental defects). The technique aligns phenotypes across organisms based on orthology (a type of homology) of genes involved in the phenotypes.
