Entrez 1854 | Ensembl ENSG00000128951 | |
 | ||
Aliases DUT, dUTPase, deoxyuridine triphosphatase External IDs MGI: 1346051 HomoloGene: 31475 GeneCards: DUT | ||
DUTP pyrophosphatase, also known as DUT, is an enzyme which in humans is encoded by the DUT gene on chromosome 15.
Contents
This gene encodes an essential enzyme of nucleotide metabolism. The encoded protein forms a ubiquitous, homotrimeric enzyme that hydrolyzes dUTP to dUMP and pyrophosphate. This reaction serves two cellular purposes: providing a precursor (dUMP) for the synthesis of thymine nucleotides needed for DNA replication, and limiting intracellular pools of dUTP. Elevated levels of dUTP lead to increased incorporation of uracil into DNA, which induces extensive excision repair mediated by uracil glycosylase. This repair process, resulting in the removal and reincorporation of dUTP, is self-defeating and leads to DNA fragmentation and cell death. Alternative splicing of this gene leads to different isoforms that localize to either the mitochondrion or nucleus. A related pseudogene is located on chromosome 19.
Structure
In humans, this gene encodes a homotrimeric enzyme with two isoforms characterized by their distinct subcellular localizations: the nuclear isoform (DUT-N) and mitochondrial isoform (DUT-M).
Gene
Northern blot analysis reveals distinct mRNA transcripts for DUT-N (1.1 kb) and DUT-M (1.4 kb). The isoforms are produced from alternative splicing at different 5' exons, with the first exon of DUT-N occurring 767 base pairs downstream of the first exon in DUT-M. Regulation at different promoters has been proposed to account for the differential expression of these isoforms.
Protein
The mature forms of DUT-N (22 kDa) and DUT-M (23 kDa) are nearly identical except for a short N-terminal region present in DUT-M. The DUT-M precursor (31 kDa) contains an arginine-rich, 69-residue mitochondrial targeting sequence which undergoes post-translational cleavage to effect mitochondrial import. Meanwhile, the monopartite NLS sequence is critical for the function and nuclear localization of DUT-N, which would otherwise accumulate in the cytoplasm. Though both isoforms contain the NLS, the sequence in DUT-M is sequestered away from cognate karyopherins. The isoelectric points of DUT-N (6.0) and DUT-M (8.1) correspond to the pH of their respective subcellular compartments.
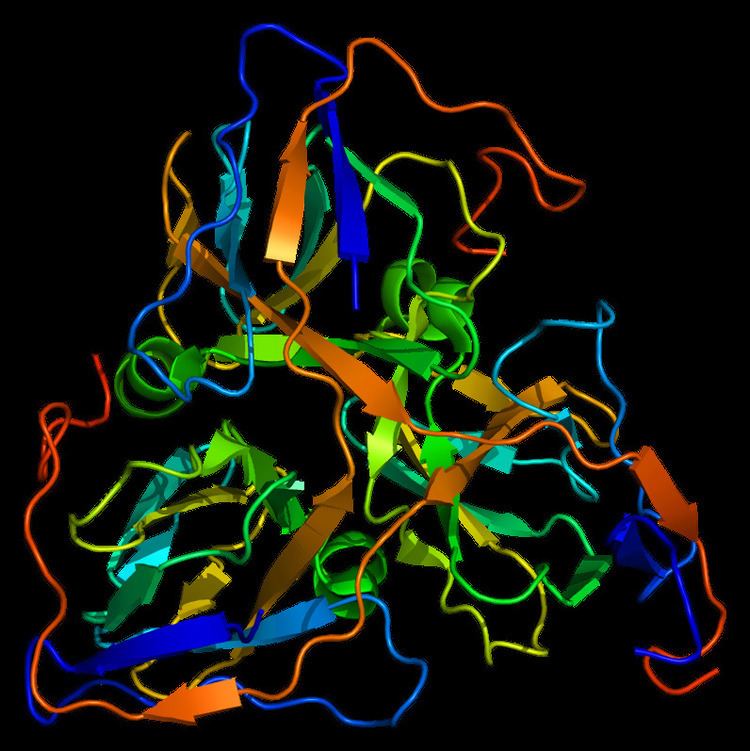
DUT is a homotrimer with three active sites formed by each of its three subunits. Typically, each subunit forms an eight-stranded barrel that swaps C-terminal β-strands with the other subunits to assemble into the trimer structure. In addition to the β-strand swapping, these subunits interact via extended bimolecular interfaces and three-fold central channel. As a member of the dUTPase family, DUT requires the presence of a divalent metal ion such as Mg2+ for their enzymatic function. DUT-N also contains a consensus cyclin-dependent kinase phosphorylation site that is phosphorylated at the serine as part of its cell cycle regulation.
Function
DUT is a member of the dUTPase family, which is known for catalyzing the pyrophosphoralysis of dUTP into dUMP and inorganic pyrophosphate. This function contributes to DNA replication and repair via de novo thymidylate biosynthesis, as the dUMP product is methylated by thymidylate synthase (TS) to form dTMP, which is then phosphorylated to dTTP. DUT is also crucial for maintaining genome integrity by reducing cellular dUTP levels, thereby preventing the repeated cycles of uracil misincorporation into DNA and DNA repair-mediated strand breaks that would lead to cell death.
In addition to their different localizations, the two DUT isoforms display different expression patterns: while DUT-M is constitutively expressed, DUT-N is under cell cycle control and notably upregulated during S phase. These expression patterns correspond with their roles in the DNA replication cycle of their respective genomes, and thus indicate different regulatory mechanisms affecting each isoform.
Mechanism
The dUTP hydrolysis cycle can be outlined in the following four enzymatic steps: (i) fast substrate binding, (ii) isomerization of the enzyme-substrate complex into the catalytically competent conformation, (iii) hydrolysis of the substrate, and (iv) rapid, non-ordered release of the products.
Clinical significance
Since many chemotherapeutic agents such as 5-fluorouracil treat neoplastic diseases, including head and neck cancer, breast cancer, and gastrointestinal cancer, by targeting TS in thymidylate metabolism, DUT may protect against the cytotoxic side effects by countering dUTP accumulation. At the same time, high levels of DUT-N have been associated with chemoresistance and faster tumor progression, and thus, could also serve as a prognostic marker for overall survival and response to chemotherapy. Similarly, DUT is significantly overexpressed in hepatocellular carcinoma and may serve as a prognostic marker for the cancer. Notably, DUT expression is regulated by the tumor suppressor gene p53 in order to promote apoptosis of tumor cells.
Interactions
DUT interacts with dUTP to catalyze its hydrolysis into dUMP and pyrophosphate. E2F and Sp1 enhance DUT expression by binding its promoter, while p53 inhibits DUT transcription by binding its promoter. A putative NF-κB binding site was also identified in the DUT promoter.
