Symbol ClpS InterPro IPR003769 SUPERFAMILY 1mbx | Pfam PF02617 SCOP 1mbx Pfam structures | |
 | ||
ClpS is an N-recognin in the N-end rule pathway. ClpS interacts with protein substrates that have a bulky hydrophobic residue (leucine, phenylalanine, tyrosine, and tryptophan) at the N-terminus. The protein substrate is then degraded by the ClpAP protease.
In molecular biology, the ATP-dependent Clp protease adaptor protein ClpS is a bacterial protein. In the bacterial cytosol, ATP-dependent protein degradation is performed by several different chaperone-protease pairs, including ClpAP. ClpS directly influences the ClpAP machine by binding to the N-terminal domain of the chaperone ClpA. The degradation of ClpAP substrates, both SsrA-tagged proteins and ClpA itself, is specifically inhibited by ClpS. ClpS modifies ClpA substrate specificity, potentially redirecting degradation by ClpAP toward aggregated proteins.
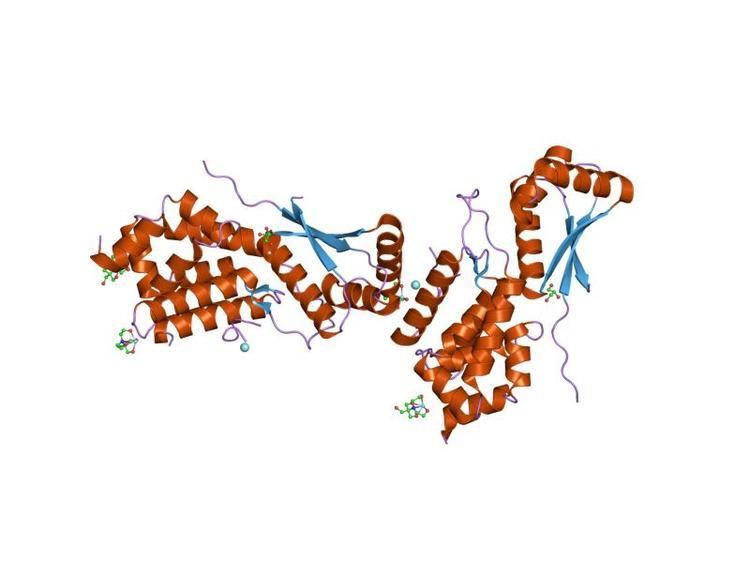
ClpS is a small alpha/beta protein that consists of three alpha-helices connected to three antiparallel beta-strands. The protein has a globular shape, with a curved layer of three antiparallel alpha-helices over a twisted antiparallel beta-sheet. Dimerization of ClpS may occur through its N-terminal domain. This short extended N-terminal region in ClpS is followed by the central seven-residue beta-strand, which is flanked by two other beta-strands in a small beta-sheet.
