Symbol EcoRII-N Pfam clan CL0405 SCOP 1na6 | Pfam PF09217 InterPro IPR015300 SUPERFAMILY 1na6 | |
 | ||
Restriction endonuclease (REase) EcoRII (pronounced "eco R two") is an enzyme of restriction modification system (RM) naturally found in Escherichia coli, a Gram-negative bacteria. Its molecular mass is 45.2 kDa, being composed of 402 amino acids.
Contents
Mode of action
EcoRII is a bacterial Type IIE REase that interacts with two or three copies of the pseudopalindromic DNA recognition sequence 5'-CCWGG-3' (W = A or T), one being the actual target of cleavage, the other(s) serving as the allosteric activator(s). EcoRII cut target DNA sequence CCWGG generating sticky ends.
Structure
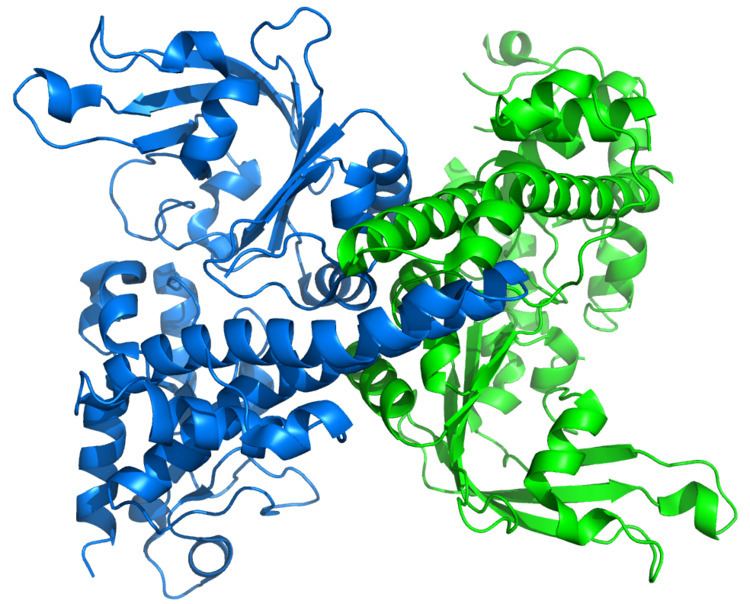
The apo crystal structure of EcoRII mutant R88A (PDB: 1NA6) has been solved at 2.1 Å resolution. The EcoRII monomer has two domains, N-terminal and C-terminal, linked through a hinge loop.
Effector-binding domain
The N-terminal effector-binding domain has an archetypal DNA-binding pseudobarrel fold (SCOP 101936) with a prominent cleft. Structural superposition showed it is evolutionarily related to:
Catalytic domain
The C-terminal catalytic domain has a typical restriction endonuclease-like fold (SCOP 52979) and belongs to the large (more than 30 members) restriction endonuclease superfamily (SCOP 52980).
Autoinhibition/activation mechanism
Structure-based sequence alignment and site-directed mutagenesis identified the putative PD..D/EXK active sites of the EcoRII catalytic domain dimer that in apo structure are spatially blocked by the N-terminal domains.
